Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors
- PMID: 1324435
- PMCID: PMC6159883
- DOI: 10.1038/358771a0
Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors
Abstract
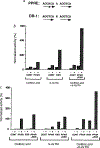
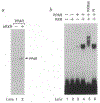
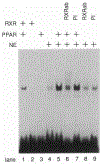
Peroxisomes are cytoplasmic organelles which are important in mammals in modulation of lipid homeostasis, including the metabolism of long-chain fatty acids and conversion of cholesterol to bile salts (reviewed in refs 1 and 2). Amphipathic carboxylates such as clofibric acid have been used in man as hypolipidaemic agents and in rodents they stimulate the proliferation of peroxisomes. These agents, termed peroxisome proliferators, and all-trans retinoic acid activate genes involved in peroxisomal-mediated beta-oxidation of fatty acids. Here we show that the receptor activated by peroxisome proliferators and the retinoid X receptor-alpha (ref. 6) form a heterodimer that activates acyl-CoA oxidase gene expression in response to either clofibric acid or the retinoid X receptor-alpha ligand, 9-cis retinoic acid, an all-trans retinoic acid metabolite; simultaneous exposure to both activators results in a synergistic induction of gene expression. These data demonstrate the coupling of the peroxisome proliferator and retinoid signalling pathways and provide evidence for a physiological role for 9-cis retinoic acid in modulating lipid metabolism.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

