The MRS1 gene of S. douglasii: co-evolution of mitochondrial introns and specific splicing proteins encoded by nuclear genes
- PMID: 1333316
- PMCID: PMC6057374
The MRS1 gene of S. douglasii: co-evolution of mitochondrial introns and specific splicing proteins encoded by nuclear genes
Abstract
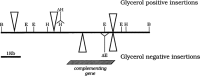
We have developed a rapid and simple methodology to locate yeast genes within cloned inserts, obtain partial sequence information, and construct chromosomal disruptions of these genes. This methodology has been used to study a nuclear gene from the yeast S. douglasii (a close relative of S. cerevisiae), which is essential for the excision of the mitochondrial intron aI1 of S. douglasii (the first intron in the gene encoding subunit I of cytochrome oxidase), an intron which is not present in the mitochondrial genome of S. cerevisiae. We have shown that this gene is the homologue of the S. cerevisiae MRS1 gene, which is essential for the excision of the mitochondrial introns bI3 and aI5 beta of S. cerevisiae, but is unable to assure the excision of the intron aI1 from the coxI gene of S. douglasii. The two genes are very similar, with only 13% nucleotide substitutions in the coding region, transitions being 2.5 times more frequent than transvertions. At the protein level there are 86% identical residues and 7% conservative substitutions. The divergence of the MRS1 genes of S. cerevisiae and S. douglasii, and the concomitant changes in the structure of their mitochondrial genomes is an interesting example of the co-evolution of nuclear and mitochondrial genomes.
Figures




References
-
- Banroques J., Delahodde A., and Jacq C. (1986), Cell 46, 837–844. - PubMed
-
- Bousquet I., Dujardin G., Poyton R. O., and Slonimski P. P. (1990), Curr Genet 18, 117–124. - PubMed
-
- Brown W. M., Prager E. M., Wang A., and Wilson A. C. (1982), J Mol Evol 18, 225–239. - PubMed
-
- Cech T. R. (1990), Annu Rev Biochem 59, 543–568. - PubMed
-
- Claisse M., Michel F., and Hawthorne D. (1987), in Plant Mitochondria. Plenum Press, London, pp. 283–292.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
