Analysis of the promoter of the cytochrome P-450 2B2 gene in the rat
- PMID: 1335316
- PMCID: PMC6057372
Analysis of the promoter of the cytochrome P-450 2B2 gene in the rat
Abstract
About 3 kb of the promoter region of the gene encoding cytochrome P-450 2B2 (CYP2B2) in the rat were sequenced and searched for potential cis-acting elements. Apart from putative binding sites for (liver-specific) protein factors, a region showing homology with the LINE 1 retrotransposon element was also found. Three proximal promoter fragments, encompassing nucleotides -579 to -372, -372 to -211, and -211 to +1, respectively, were shown to contain binding sites for multiple protein factors by bandshift analyses. The strongest protein-binding element, designated BRE (basic regulatory element), occurs between -103 to -66. Its structure is very similar to a negative control element in the murine cmyc promoter and displays a composite feature having a tandemly repeated sequence homology with the BTE (basic transcription element; Yanagida et al., 1990) separated by a CCAAA-box. The use of a deletion series of this template in in vitro transcription assays, provided evidence that the BRE serves as a major cis-acting element in the (regulated) transcription activation of the CYP2B2 gene.
Figures

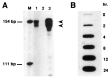
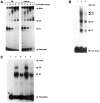
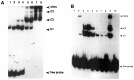



References
-
- Cereghini S., Raymondjean M., Carranca A. G., Herbomel M., and Yaniv M. (1987), Cell 50, 627–638. - PubMed
-
- Chomczynski P. and Sacchi N. (1987) Anal Biochem 64, 156–159. - PubMed
-
- Corcoran L. M., Cory S., and Adams J. M. (1985), Cell 40, 71–79. - PubMed
-
- Denison M. S., Fisher J. M., and Whitlock J. P. Jr. (1988), J Biol Chem 263, 17221–17224. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
