Mouse F9 teratocarcinoma stem cells expressing the stably transfected homeobox gene Hox 1.6 exhibit an altered morphology
- PMID: 1353003
- PMCID: PMC6057386
Mouse F9 teratocarcinoma stem cells expressing the stably transfected homeobox gene Hox 1.6 exhibit an altered morphology
Abstract
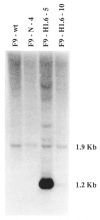
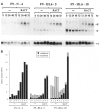
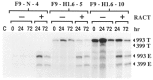
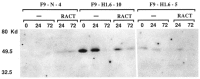
Expression of the murine homeobox gene Hox 1.6 rapidly increases in F9 teratocarcinoma cells when these cells are induced with retinoic acid to differentiate into primitive and parietal endoderm. Hox 1.6 encodes a putative transcriptional regulatory protein which may function as a secondary regulator of gene expression during the differentiation process. To examine the role of the Hox 1.6 gene, we have stably transfected F9 stem cells with a cDNA containing the complete coding sequence of Hox 1.6 under the control of the mouse metallothionein promoter. Two clonally distinct cell lines that express high levels of the transfected Hox 1.6 gene have been isolated and characterized. We show that expression of the transfected Hox 1.6 gene in F9 cells dramatically alters the stem cell morphology. However, the transfected cells do not differentiate in the absence of retinoic acid treatment, nor are they prevented from differentiating in response to such treatments. We therefore suggest that the Hox 1.6 gene controls the expression of genes which influence changes in F9 cell morphology during RA-induced differentiation.
Figures



References
-
- Ausebel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., and Struhl K., eds. (1987), Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York.
-
- Berget S. M. and Sharp P. A. (1979), J Mol Biol 129, 547–565. - PubMed
-
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., and Rutter W. J. (1979), Biochemistry 18, 5294–5299. - PubMed
-
- Chisaka O., Musci T. S., and Capecchi M. R. (1992), Nature 355, 516–520. - PubMed
-
- Colberg-Poley A., Voss S., Chowdhury K., and Gruss P. (1985), Nature 314, 731–738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
