Ordering gene function: the interpretation of epistasis in regulatory hierarchies
- PMID: 1365397
- PMCID: PMC3955268
- DOI: 10.1016/0168-9525(92)90263-4
Ordering gene function: the interpretation of epistasis in regulatory hierarchies
Abstract
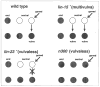
The order of action of genes in a regulatory hierarchy that is governed by a signal can often be determined by the method of epistasis analysis, in which the phenotype of a double mutant is compared with that of single mutants. The epistatic mutation may be in either the upstream or the downstream gene, depending on the nature of the two mutations and the type of regulation. Nevertheless, when the regulatory hierarchy satisfies certain conditions, simple rules allow the position of the epistatic locus in the pathway to be determined without detailed knowledge of the nature of the mutations, the pathway, or the molecular mechanism of regulation.
Figures


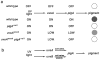
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

