Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals
- PMID: 13678422
- PMCID: PMC222960
- DOI: 10.1186/1471-2105-4-42
Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals
Abstract
Background: The kelch motif is an ancient and evolutionarily-widespread sequence motif of 44-56 amino acids in length. It occurs as five to seven repeats that form a beta-propeller tertiary structure. Over 28 kelch-repeat proteins have been sequenced and functionally characterised from diverse organisms spanning from viruses, plants and fungi to mammals and it is evident from expressed sequence tag, domain and genome databases that many additional hypothetical proteins contain kelch-repeats. In general, kelch-repeat beta-propellers are involved in protein-protein interactions, however the modest sequence identity between kelch motifs, the diversity of domain architectures, and the partial information on this protein family in any single species, all present difficulties to developing a coherent view of the kelch-repeat domain and the kelch-repeat protein superfamily. To understand the complexity of this superfamily of proteins, we have analysed by bioinformatics the complement of kelch-repeat proteins encoded in the human genome and have made comparisons to the kelch-repeat proteins encoded in other sequenced genomes.
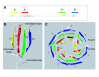
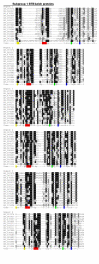
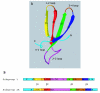
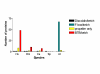
Results: We identified 71 kelch-repeat proteins encoded in the human genome, whereas 5 or 8 members were identified in yeasts and around 18 in C. elegans, D. melanogaster and A. gambiae. Multiple domain architectures were identified in each organism, including previously unrecognised forms. The vast majority of kelch-repeat domains are predicted to form six-bladed beta-propellers. The most prevalent domain architecture in the metazoan animal genomes studied was the BTB/kelch domain organisation and we uncovered 3 subgroups of human BTB/kelch proteins. Sequence analysis of the kelch-repeat domains of the most robustly-related subgroups identified differences in beta-propeller organisation that could provide direction for experimental study of protein-binding characteristics.
Conclusion: The kelch-repeat superfamily constitutes a distinct and evolutionarily-widespread family of beta-propeller domain-containing proteins. Expansion of the family during the evolution of multicellular animals is mainly accounted for by a major expansion of the BTB/kelch domain architecture. BTB/kelch proteins constitute 72 % of the kelch-repeat superfamily of H. sapiens and form three subgroups, one of which appears the most-conserved during evolution. Distinctions in propeller blade organisation between subgroups 1 and 2 were identified that could provide new direction for biochemical and functional studies of novel kelch-repeat proteins.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

