Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II
- PMID: 1371280
- PMCID: PMC3373963
Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II
Abstract
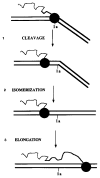
In addition to polynucleotide polymerization, DNA polymerases and bacterial RNA polymerase can also remove nucleotides from the growing end of nucleic acid chains. For DNA polymerases this activity is an important factor in establishing fidelity in DNA synthesis. This report describes a novel in vitro activity of RNA polymerase II whereby it cleaves an RNA chain contained within an active elongation complex. These elongation complexes are arrested at a previously identified, naturally occurring transcriptional pause site in a human gene. The new 3'-end revealed by this cleavage remains associated with an active elongation complex and is capable of being extended by RNA polymerase II. Nascent RNA cleavage is evident after removal of free nucleotides and is dependent upon a divalent metal cation and transcription elongation factor SII. This function of SII could be important in its function as an activator of transcription elongation. It is also possible that the transcript cleavage activity of RNA polymerase II represents a proofreading function of the enzyme.
Figures
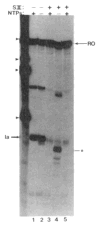

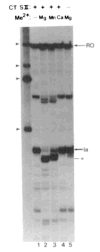
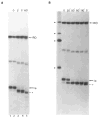
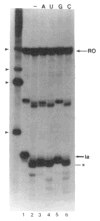
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

