The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage
- PMID: 1379232
- PMCID: PMC3371615
The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage
Abstract
Regulation of transcription elongation is an important mechanism in controlling eukaryotic gene expression. SII is an RNA polymerase II-binding protein that stimulates transcription elongation and also activates nascent transcript cleavage by RNA polymerase II in elongation complexes in vitro (Reines, D. (1992) J. Biol. Chem. 267, 3795-3800). Here we show that SII-dependent in vitro transcription through an arrest site in a human gene is preceded by nascent transcript cleavage. RNA cleavage appeared to be an obligatory step in the SII activation process. Recombinant SII activated cleavage while a truncated derivative lacking polymerase binding activity did not. Cleavage was not restricted to an elongation complex arrested at this particular site, showing that nascent RNA hydrolysis is a general property of RNA polymerase II elongation complexes. These data support a model whereby SII stimulates elongation via a ribonuclease activity of the elongation complex.
Figures

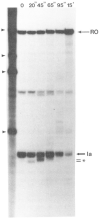
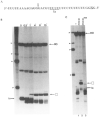
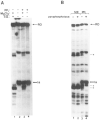
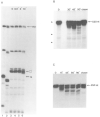
References
-
- Agarwal K, Baek K, Jeon C, Miyamoto K, Ueno A, Yoon H. Biochemistry. 1991;30:7842–7851. - PubMed
-
- Arndt KM, Chamberlin MJ. J. Mol. Biol. 1990;213:79–108. - PubMed
-
- Atkinson MR, Deutscher MP, Kornberg A, Russell AF, Moffatt JG. Biochemistry. 1969;8:4897–4904. - PubMed
-
- Chafin DR, Claussen TJ, Price DH. J. Biol. Chem. 1991;266:9256–9262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

