A novel design of whole-genome microarray probes for Saccharomyces cerevisiae which minimizes cross-hybridization
- PMID: 14499002
- PMCID: PMC239980
- DOI: 10.1186/1471-2164-4-38
A novel design of whole-genome microarray probes for Saccharomyces cerevisiae which minimizes cross-hybridization
Abstract
Background: Numerous DNA microarray hybridization experiments have been performed in yeast over the last years using either synthetic oligonucleotides or PCR-amplified coding sequences as probes. The design and quality of the microarray probes are of critical importance for hybridization experiments as well as subsequent analysis of the data.
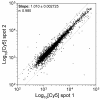
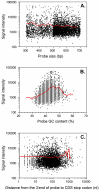
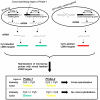
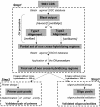
Results: We present here a novel design of Saccharomyces cerevisiae microarrays based on a refined annotation of the genome and with the aim of reducing cross-hybridization between related sequences. An effort was made to design probes of similar lengths, preferably located in the 3'-end of reading frames. The sequence of each gene was compared against the entire yeast genome and optimal sub-segments giving no predicted cross-hybridization were selected. A total of 5660 novel probes (more than 97% of the yeast genes) were designed. For the remaining 143 genes, cross-hybridization was unavoidable. Using a set of 18 deletant strains, we have experimentally validated our cross-hybridization procedure. Sensitivity, reproducibility and dynamic range of these new microarrays have been measured. Based on this experience, we have written a novel program to design long oligonucleotides for microarray hybridizations of complete genome sequences.
Conclusions: A validated procedure to predict cross-hybridization in microarray probe design was defined in this work. Subsequently, a novel Saccharomyces cerevisiae microarray (which minimizes cross-hybridization) was designed and constructed. Arrays are available at Eurogentec S. A. Finally, we propose a novel design program, OliD, which allows automatic oligonucleotide design for microarrays. The OliD program is available from authors.
Figures


References
-
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

