Positive and negative cis-acting regulatory sequences control expression of leukotoxin in Actinobacillus actinomycetemcomitans 652
- PMID: 14500484
- PMCID: PMC201044
- DOI: 10.1128/IAI.71.10.5640-5649.2003
Positive and negative cis-acting regulatory sequences control expression of leukotoxin in Actinobacillus actinomycetemcomitans 652
Abstract
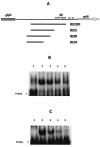
Integration of IS1301 into an AT-rich inverted repeat located upstream of the ltx operon was previously shown to confer a hyperleukotoxic phenotype in Actinobacillus actinomycetemcomitans IS1 (T. He, T. Nishihara, D. R. Demuth, and I. Ishikawa, J. Periodontol. 70:1261-1268, 1999), but the mechanism leading to increased leukotoxin production was not determined. We show that an IS1 ltx promoter::lacZ reporter construct expresses 12-fold higher levels of beta-galactosidase activity than a reporter containing the ltx promoter from A. actinomycetemcomitans 652, suggesting that IS1301 increases transcription of the ltx operon. Examination of the IS1301 sequence identified a potential outwardly directed promoter. However, site-specific mutagenesis of the -35 element of the putative promoter had no effect on the transcriptional activity of the IS1 reporter construct. Furthermore, reverse transcriptase PCR and real-time PCR experiments did not detect a transcript that was initiated within IS1301. These results suggest that increased expression of leukotoxin in strain IS1 does not arise from an outwardly directed IS1301 promoter. To determine how IS1301 alters transcriptional regulation of the ltx operon, cis-acting sequences that regulate leukotoxin expression were identified. The AT-rich sequence that resides downstream from the site of IS1301 insertion was shown to function as a positive cis-acting regulator of leukotoxin expression. This sequence resembles an UP element in its location, AT-rich content, and activity and is homologous to the consensus UP element sequence. In addition, a negative cis-acting sequence was identified upstream from the site of IS1301 insertion, and deletion of this region increased promoter activity by fourfold. Mobility shift experiments showed that this region bound to a protein(s) in extracts from A. actinomycetemcomitans 652. The specific sequences required for this interaction were localized to a 26-nucleotide segment of the ltx promoter that resides 17 bp upstream from the site of IS1301 insertion. Together, these results suggest that positive and negative cis-acting sequences regulate leukotoxin expression and that IS1301 may increase transcription of the ltx operon in A. actinomycetemcomitans IS1 by displacing a negative cis-acting regulator approximately 900 bp upstream from the basal elements of the ltx promoter.
Figures




References
-
- Block, P. J., A. C. Fox, C. Yoran, and A. J. Kaltman. 1973. Actinobacillus actinomycetemcomitans endocarditis: report of a case and review of the literature. Am. J. Med. Sci. 276:387-392. - PubMed
-
- Brogan, J. M., E. T. Lally, and D. R. Demuth. 1996. Construction of pYGK, an Actinobacillus actinomycetemcomitans/Escherichia coli shuttle vector. Gene 169:141-142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

