Acid-adaptive genes of Helicobacter pylori
- PMID: 14500513
- PMCID: PMC201084
- DOI: 10.1128/IAI.71.10.5921-5939.2003
Acid-adaptive genes of Helicobacter pylori
Abstract
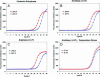
Helicobacter pylori is the only neutralophile that has been able to colonize the human stomach by using a variety of acid-adaptive mechanisms. One of the adaptive mechanisms is increased buffering due to expression of an acid-activated inner membrane urea channel, UreI, and a neutral pH-optimum intrabacterial urease. To delineate other possible adaptive mechanisms, changes in gene expression in response to acid exposure were examined using genomic microarrays of H. pylori exposed to different levels of external pH (7.4, 6.2, 5.5, and 4.5) for 30 min in the absence and presence of 5 mM urea. Gene expression was correlated with intrabacterial pH measured using 2',7'-bis-(2-carboxyethyl)-5-carboxyfluorescein and compared to that observed with exposure to 42 degrees C for 30 min. Microarrays containing the 1,534 open reading frames of H. pylori strain 26695 were hybridized with cDNAs from control (pH 7.4; labeled with Cy3) and acidic (labeled with Cy5) conditions. The intrabacterial pH was 8.1 at pH 7.4, fell to 5.3 at pH 4.5, and rose to 6.2 with urea. About 200 genes were up-regulated and approximately 100 genes were down-regulated at pH 4.5 in the absence of urea, and about half that number changed in the presence of urea. These genes included pH-homeostatic, transcriptional regulatory, motility, cell envelope, and pathogenicity genes. The up-regulation of some pH-homeostatic genes was confirmed by real-time PCR. There was little overlap with the genes induced by temperature stress. These results suggest that H. pylori has evolved multifaceted acid-adaptive mechanisms enabling it to colonize the stomach that may be novel targets for eliminating infection.
Figures



References
-
- Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. - PubMed
-
- Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

