Analysis of expression profile of mammalian cell entry (mce) operons of Mycobacterium tuberculosis
- PMID: 14500535
- PMCID: PMC201107
- DOI: 10.1128/IAI.71.10.6083-6087.2003
Analysis of expression profile of mammalian cell entry (mce) operons of Mycobacterium tuberculosis
Abstract
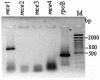
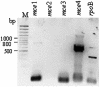
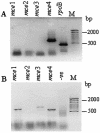
The sequencing of the complete genome of M. tuberculosis H37Rv has resulted in the recognition of four mce operons in its genome by in silico analysis. In an attempt to understand the significance of the redundancy of mce operons, we analyzed the expression profile of mce operons after different periods of growth in culture as well as during in vivo infection. Our results strongly suggest that mce1 is expressed as a polycistronic message. In culture from day 8 to day 12, expression of only mce1 was observed, but as the cultures progress towards stationary phase the expression profile of mce operons was altered; the transcripts of the mce1 operon were barely detected while those of the mce4 operon were prominent. In an analysis of the expression of mce operons in tubercle material collected from infected animal tissues, we detected the expression of mce1, -3 and -4. Our results imply that mce operons other than mce1 are also expressed during infection and that it is necessary to examine their role in pathogenesis.
Figures


References
-
- Arruda, S., G. Bomfim, R. Knights, Huima-Byron, and L. W. Riley. 1993. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261:1454-1457. - PubMed
-
- Chitale, S., S. Ehrt, S. Kawamura, T. Fujimura, N. Shimono, N. Anand, S. Lu, L. Cohen-Gould, and L. W. Riley. 2001. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 3:247-254. - PubMed
-
- Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, S. V. Harris, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

