Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis
- PMID: 14500781
- PMCID: PMC208781
- DOI: 10.1073/pnas.1634991100
Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis
Abstract
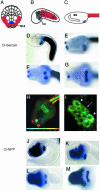
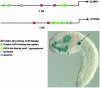
Here we exploit the extensive cell lineage information and streamlined genome of the ascidian, Ciona intestinalis, to investigate heart development in a basal chordate. Several cardiac genes were analyzed, including the sole Ciona ortholog of the Drosophila tinman gene, and tissue-specific enhancers were isolated for some of the genes. Conserved sequence motifs within these enhancers facilitated the isolation of a heart enhancer for the Ciona Hand-like gene. Altogether, these studies provide a regulatory framework for the differentiation of the cardiac mesoderm, beginning at the 110-cell stage, and extending through the fusion of cardiac progenitors during tail elongation. The cardiac lineage shares a common origin with the germ line, and zygotic transcription is first detected in the heart progenitors only after its separation from the germ line at the 64-cell stage. We propose that germ-line determinants influence the specification of the cardiac mesoderm, both by inhibiting inductive signals required for the development of noncardiac mesoderm lineages, and by providing a localized source of Wnt-5 and other signals required for heart development. We discuss the possibility that the germ line also influences the specification of the vertebrate heart.
Figures



References
-
- Andree, B., Duprez, D., Vorbusch, B., Arnold, H. H. & Brand, T. (1998) Mech. Dev. 70, 119–131. - PubMed
-
- Cripps, R. M. & Olson, E. N. (2002) Dev. Biol. 246, 14–28. - PubMed
-
- Pandur, P., Lasche, M., Eisenberg, L. M. & Kuhl, M. (2002) Nature 418, 636–641. - PubMed
-
- Reifers, F., Walsh, E. C., Leger, S., Stainier, D. Y. & Brand, M. (2000) Development (Cambridge, U.K.) 127, 225–235. - PubMed
-
- Satoh, N. (1994) Developmental Biology of Ascidians (Cambridge Univ. Press, New York).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

