GLIS3, a novel member of the GLIS subfamily of Krüppel-like zinc finger proteins with repressor and activation functions
- PMID: 14500813
- PMCID: PMC206473
- DOI: 10.1093/nar/gkg776
GLIS3, a novel member of the GLIS subfamily of Krüppel-like zinc finger proteins with repressor and activation functions
Abstract
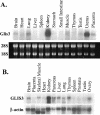
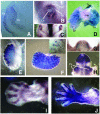
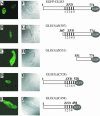
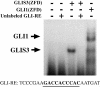
In this study, we describe the identification and characterization of a novel transcription factor GLI-similar 3 (GLIS3). GLIS3 is an 83.8 kDa nuclear protein containing five C2H2-type Krüppel-like zinc finger motifs that exhibit 93% identity with those of GLIS1, however, little homology exists outside their zinc finger domains. GLIS3 can function as a repressor and activator of transcription. Deletion mutant analysis determined that the N- and C-termini are required for optimal transcriptional activity. GLIS3 binds to the GLI-RE consensus sequence and is able to enhance GLI-RE-dependent transcription. GLIS3(DeltaC496), a dominant-negative mutant, inhibits transcriptional activation by GLIS3 and GLI1. Whole mount in situ hybridization on mouse embryos from stage E6.5 through E14.5 demonstrated that GLIS3 is expressed in specific regions in developing kidney and testis and in a highly dynamic pattern during neurulation. From E11.5 through E12.5 GLIS3 was strongly expressed in the interdigital regions, which are fated to undergo apoptosis. The temporal and spatial pattern of GLIS3 expression observed during embryonic development suggests that it may play a critical role in the regulation of a variety of cellular processes during development. Both the repressor and activation functions of GLIS3 may be involved in this control.
Figures











References
-
- Nagai T., Aruga,J., Takada,S., Gunther,T., Sporle,R., Schughart,K. and Mikoshiba,K. (1997) The expression of the mouse Zic1, Zic2 and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol., 182, 299–313. - PubMed
-
- Aruga J., Nagai,T., Tokuyama,T., Hayashizaki,Y., Okazaki,Y., Chapman,V.M. and Mikoshiba,K. (1996) The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J. Biol. Chem., 271, 1043–1047. - PubMed
-
- Kinzler K.W., Ruppert,J.M., Bigner,S.H. and Vogelstein,B. (1988) The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature, 332, 371–374. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

