A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage
- PMID: 14500814
- PMCID: PMC206461
- DOI: 10.1093/nar/gkg761
A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage
Abstract
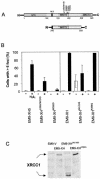
The molecular role of poly (ADP-ribose) polymerase-1 in DNA repair is unclear. Here, we show that the single-strand break repair protein XRCC1 is rapidly assembled into discrete nuclear foci after oxidative DNA damage at sites of poly (ADP-ribose) synthesis. Poly (ADP-ribose) synthesis peaks during a 10 min treatment with H2O2 and the appearance of XRCC1 foci peaks shortly afterwards. Both sites of poly (ADP-ribose) and XRCC1 foci decrease to background levels during subsequent incubation in drug-free medium, consistent with the rapidity of the single-strand break repair process. The formation of XRCC1 foci at sites of poly (ADP-ribose) was greatly reduced by mutation of the XRCC1 BRCT I domain that physically interacts with PARP-1. Moreover, we failed to detect XRCC1 foci in Adprt1-/- MEFs after treatment with H2O2. These data demonstrate that PARP-1 is required for the assembly or stability of XRCC1 nuclear foci after oxidative DNA damage and suggest that the formation of these foci is mediated via interaction with poly (ADP-ribose). These results support a model in which the rapid activation of PARP-1 at sites of DNA strand breakage facilitates DNA repair by recruiting the molecular scaffold protein, XRCC1.
Figures





References
-
- Xu Y.J., Kim,E.Y. and Demple,B. (1998) Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J. Biol. Chem., 273, 28837–28844. - PubMed
-
- Caldecott K.W. (2001) Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays, 23, 447–455. - PubMed
-
- Ochs K., Sobol,R.W., Wilson,S.H. and Kaina,B. (1999) Cells deficient in DNA polymerase beta are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Res., 59, 1544–1551. - PubMed
-
- Thompson L.H., Brookman,K.W., Dillehay,L.E., Carrano,A.V., Mazrimas,J.A., Mooney,C.L. and Minkler,J.L. (1982) A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res., 95, 427–440. - PubMed
-
- Dominguez I., Daza,P., Natarajan,A.T. and Cortes,F. (1998) A high yield of translocations parallels the high yield of sister chromatid exchanges in the CHO mutant EM9. Mutat. Res., 398, 67–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

