Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences
- PMID: 14500827
- PMCID: PMC206480
- DOI: 10.1093/nar/gkg786
Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences
Abstract
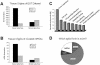
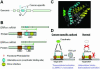
We report here a genome-wide analysis of alternative splicing in 2 million human expressed sequence tags (ESTs), to identify splice forms that are up-regulated in tumors relative to normal tissues. We found strong evidence (P < 0.01) of cancer-specific splice variants in 316 human genes. In total, 78% of the cancer-specific splice forms we detected are confirmed by human-curated mRNA sequences, indicating that our results are not due to random mis-splicing in tumors; 73% of the genes showed the same cancer-specific splicing changes in tissue-matched tumor versus normal datasets, indicating that the vast majority of these changes are associated with tumorigenesis, not tissue specificity. We have confirmed our EST results in an independent set of experimental data provided by human-curated mRNAs (P-value 10(-5.7)). Moreover, the majority of the genes we detected have functions associated with cancer (P-value 0.0007), suggesting that their altered splicing may play a functional role in cancer. Analysis of the types of cancer-specific splicing shifts suggests that many of these shifts act by disrupting a tumor suppressor function. Sur prisingly, our data show that for a large number (190 in this study) of cancer-associated genes cloned originally from tumors, there exists a previously uncharacterized splice form of the gene that appears to be predominant in normal tissue.
Figures

References
-
- Ross D.T., Scherf,U., Eisen,M.B., Perou,C.M., Rees,C., Spellman,P., Iyer,V., Jeffrey,S.S., Van de Rijn,M., Waltham,M. et al. (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet., 24, 227–235. - PubMed
-
- Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. - PubMed
-
- Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. - PubMed
-
- IHGS Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

