A microarray-based high throughput molecular marker genotyping method: the tagged microarray marker (TAM) approach
- PMID: 14500843
- PMCID: PMC206481
- DOI: 10.1093/nar/gng113
A microarray-based high throughput molecular marker genotyping method: the tagged microarray marker (TAM) approach
Abstract
A microarray-based method has been developed for scoring thousands of DNAs for a co-dominant molecular marker on a glass slide. The approach was developed to detect insertional polymorphism of transposons and works well with single nucleotide polymorphism (SNP) markers. Biotin- terminated allele-specific PCR products are spotted unpurified onto streptavidin-coated glass slides and visualised by hybridisation of fluorescent detector oligonucleotides to tags attached to the allele- specific PCR primers. Two tagged primer oligonucleotides are used per locus and each tag is detected by hybridisation to a concatameric DNA probe labelled with multiple fluorochromes.
Figures
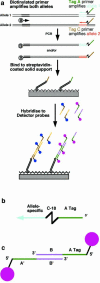
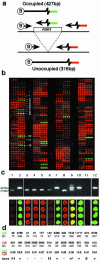



References
-
- Jeffreys A., Wilson,V. and Thein,S.L. (1985) Hypervariable ‘minisatellite’ regions in human DNA. Nature, 314, 67–73. - PubMed
-
- Tsuchihashi Z. and Dracopoli,N.C. (2002) Progress in high throughput SNP genotyping methods. Pharmacogenomics J., 2, 103–110. - PubMed
-
- Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. - PubMed

