PNA microarrays for hybridisation of unlabelled DNA samples
- PMID: 14500847
- PMCID: PMC206485
- DOI: 10.1093/nar/gng120
PNA microarrays for hybridisation of unlabelled DNA samples
Abstract
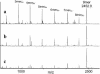
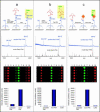
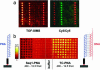
Several strategies have been developed for the production of peptide nucleic acid (PNA) microarrays by parallel probe synthesis and selective coupling of full-length molecules. Such microarrays were used for direct detection of the hybridisation of unlabelled DNA by time-of-flight secondary ion mass spectrometry. PNAs were synthesised by an automated process on filter-bottom microtitre plates. The resulting molecules were released from the solid support and attached without any purification to microarray surfaces via the terminal amino group itself or via modifications, which had been chemically introduced during synthesis. Thus, only full-length PNA oligomers were attached whereas truncated molecules, produced during synthesis because of incomplete condensation reactions, did not bind. Different surface chemistries and fitting modifications of the PNA terminus were tested. For an examination of coupling selectivity, bound PNAs were cleaved off microarray surfaces and analysed by MALDI-TOF mass spectrometry. Additionally, hybridisation experiments were performed to compare the attachment chemistries, with fully acetylated PNAs spotted as controls. Upon hybridisation of unlabelled DNA to such microarrays, binding events could be detected by visualisation of phosphates, which are an integral part of nucleic acids but missing entirely in PNA probes. Overall best results in terms of selectivity and sensitivity were obtained with thiol-modified PNAs on maleimide surfaces.
Figures



References
-
- Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. - PubMed
-
- Dawson E., Abecasis,G.R., Bumpstead,S., Chen,Y., Hunt,S., Beare,D.M., Pabial,J., Dibling,T., Tinsley,E., Kirby,S. et al. (2002) A first-generation linkage disequilibrium map of human chromosome 22. Nature, 418, 544–548. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

