Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy
- PMID: 14504273
- PMCID: PMC1705954
- DOI: 10.1074/jbc.M309238200
Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy
Abstract
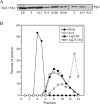
Cells must regulate both biosynthesis and degradation to ensure proper homeostasis of cellular organelles and proteins. This balance is demonstrated in a unique way in the yeast Saccharomyces cerevisiae, which possesses two distinct, yet mechanistically related trafficking routes mediating the delivery of proteins from the cytoplasm to the vacuole: the biosynthetic cytoplasm to vacuole targeting (Cvt) and the degradative autophagy pathways. Several components employed by these two transport routes have been identified, but their mechanistic interactions remain largely unknown. Here we report a novel gene involved in these pathways, which we have named ATG23. Atg23 localizes to the pre-auto-phagosomal structure but also to other cytosolic punctate compartments. Our characterization of the Atg23 protein indicates that it is required for the Cvt pathway and efficient autophagy but not pexophagy. In the absence of Atg23, cargo molecules such as prApe1 are correctly recruited to a pre-autophagosomal structure that is unable to give rise to Cvt vesicles. We also demonstrate that Atg23 is a peripheral membrane protein that requires the presence of Atg9/Apg9 to be specifically targeted to lipid bilayers. Atg9 transiently interacts with Atg23 suggesting that it participates in the recruitment of this protein.
Figures





Similar articles
-
Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway.J Biol Chem. 2004 Jul 16;279(29):29889-94. doi: 10.1074/jbc.M404399200. Epub 2004 May 11. J Biol Chem. 2004. PMID: 15138258 Free PMC article.
-
Atg23 is a vesicle-tethering protein.Autophagy. 2022 Oct;18(10):2510-2511. doi: 10.1080/15548627.2022.2105107. Epub 2022 Aug 1. Autophagy. 2022. PMID: 35867625 Free PMC article.
-
Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways.J Biol Chem. 2001 Aug 10;276(32):30442-51. doi: 10.1074/jbc.M102342200. Epub 2001 May 29. J Biol Chem. 2001. PMID: 11382760 Free PMC article.
-
Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review.
-
Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae.Curr Opin Cell Biol. 2002 Aug;14(4):468-75. doi: 10.1016/s0955-0674(02)00343-5. Curr Opin Cell Biol. 2002. PMID: 12383798 Review.
Cited by
-
The Cytoplasm-to-Vacuole Targeting Pathway: A Historical Perspective.Int J Cell Biol. 2012;2012:142634. doi: 10.1155/2012/142634. Epub 2012 Feb 20. Int J Cell Biol. 2012. PMID: 22481942 Free PMC article.
-
Mec1-mediated Atg9 phosphorylation regulates the PAS recruitment of Atg9 vesicles upon energy stress.Proc Natl Acad Sci U S A. 2025 Feb 11;122(6):e2422582122. doi: 10.1073/pnas.2422582122. Epub 2025 Feb 6. Proc Natl Acad Sci U S A. 2025. PMID: 39913206 Free PMC article.
-
Atg27 is required for autophagy-dependent cycling of Atg9.Mol Biol Cell. 2007 Feb;18(2):581-93. doi: 10.1091/mbc.e06-07-0612. Epub 2006 Nov 29. Mol Biol Cell. 2007. PMID: 17135291 Free PMC article.
-
Current opinions on mitophagy in fungi.Autophagy. 2023 Mar;19(3):747-757. doi: 10.1080/15548627.2022.2098452. Epub 2022 Jul 11. Autophagy. 2023. PMID: 35793406 Free PMC article. Review.
-
Dimerization-dependent membrane tethering by Atg23 is essential for yeast autophagy.Cell Rep. 2022 Apr 19;39(3):110702. doi: 10.1016/j.celrep.2022.110702. Cell Rep. 2022. PMID: 35443167 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

