Exclusively targeting beta-secretase to lipid rafts by GPI-anchor addition up-regulates beta-site processing of the amyloid precursor protein
- PMID: 14504402
- PMCID: PMC208827
- DOI: 10.1073/pnas.1635130100
Exclusively targeting beta-secretase to lipid rafts by GPI-anchor addition up-regulates beta-site processing of the amyloid precursor protein
Abstract
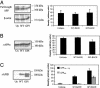
beta-Secretase (BACE, Asp-2) is a transmembrane aspartic proteinase responsible for cleaving the amyloid precursor protein (APP) to generate the soluble ectodomain sAPPbeta and its C-terminal fragment CTFbeta. CTFbeta is subsequently cleaved by gamma-secretase to produce the neurotoxic/synaptotoxic amyloid-beta peptide (Abeta) that accumulates in Alzheimer's disease. Indirect evidence has suggested that amyloidogenic APP processing may preferentially occur in lipid rafts. Here, we show that relatively little wild-type BACE is found in rafts prepared from a human neuroblastoma cell line (SH-SY5Y) by using Triton X-100 as detergent. To investigate further the significance of lipid rafts in APP processing, a glycosylphosphatidylinositol (GPI) anchor has been added to BACE, replacing the transmembrane and C-terminal domains. The GPI anchor targets the enzyme exclusively to lipid raft domains. Expression of GPIBACE substantially up-regulates the secretion of both sAPPbeta and amyloid-beta peptide over levels observed from cells overexpressing wild-type BACE. This effect was reversed when the lipid rafts were disrupted by depleting cellular cholesterol levels. These results suggest that processing of APP to the amyloid-beta peptide occurs predominantly in lipid rafts and that BACE is the rate-limiting enzyme in this process. The processing of the APP695 isoform by GPI-BACE was up-regulated 20-fold compared with wild-type BACE, whereas only a 2-fold increase in the processing of APP751/770 was seen, implying a differential compartmentation of the APP isoforms. Changes in the local membrane environment during aging may facilitate the cosegregation of APP and BACE leading to increased beta-amyloid production.
Figures






References
-
- Hussain, I., Powell, D. J., Howlett, D. R., Tew, D. G., Meek, T. D., Chapman, C., Gloger, I. S., Murphy, K. E., Southan, C., Ryan, D. M., et al. (1999) Mol. Cell. Neurosci. 14, 419–427. - PubMed
-
- Sinha, S., Anderson, J. P., Barbour, R., Basi, G. S., Caccavello, R., Davis, D., Doan, M., Dovey, H. F., Frigon, N., Hong, J., et al. (1999) Nature 402, 537–540. - PubMed
-
- Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., Teplow, D. B., Ross, S., Amarante, P., Loeloff, R., et al. (1999) Science 286, 735–741. - PubMed
-
- De Strooper, B. (2003) Neuron 38, 9–12. - PubMed
-
- Hooper, N. M. & Turner, A. J. (2002) Curr. Med. Chem. 9, 1107–1119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

