Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p
- PMID: 14507714
- PMCID: PMC1303475
- DOI: 10.1016/S0006-3495(03)74674-8
Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p
Abstract
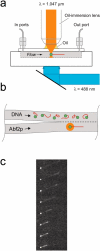
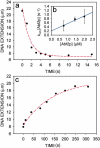
Mitochondrial and nuclear DNA are packaged by proteins in a very different manner. Although protein-DNA complexes called "nucleoids" have been identified as the genetic units of mitochondrial inheritance in yeast and man, little is known about their physical structure. The yeast mitochondrial protein Abf2p was shown to be sufficient to compact linear dsDNA, without the benefit of supercoiling, using optical and atomic force microscopy single molecule techniques. The packaging of DNA by Abf2p was observed to be very weak as evidenced by a fast Abf2p off-rate (k(off) = 0.014 +/- 0.001 s(-1)) and the extremely small forces (<0.6 pN) stabilizing the condensed protein-DNA complex. Atomic force microscopy images of individual complexes showed the 190-nm structures are loosely packaged relative to nuclear chromatin. This organization may leave mtDNA accessible for transcription and replication, while making it more vulnerable to damage.
Figures


References
-
- Brewer, L. R., M. Corzett, and R. Balhorn. 1999. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 286:120–123. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

