HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53
- PMID: 14507994
- PMCID: PMC218704
- DOI: 10.1073/pnas.2030930100
HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53
Abstract
The RING finger proteins HdmX and Hdm2 share significant structural and functional similarity. Hdm2 is a member of the RING finger family of ubiquitin-protein ligases E3 and targets the tumor suppressor protein p53 for degradation. Although HdmX also binds to p53, HdmX does not induce p53 degradation. Moreover, HdmX has been reported to interfere with p53 degradation in overexpression experiments. To obtain insight into the mechanism by which HdmX interferes with p53 degradation, we studied the effect of HdmX on the E3 activity of Hdm2 in vitro. Surprisingly, this revealed that HdmX stimulates Hdm2-mediated ubiquitination of p53 and that HdmX facilitates ubiquitination of Hdm2 and vice versa. In addition, down-regulation of HdmX expression within cells results in the accumulation of both p53 and Hdm2. Because HdmX alone does not have appreciable E3 activity, these data indicate that HdmX acts as a stimulator, rather than as an inhibitor, of the E3 activity of Hdm2 and that, at least under certain conditions, HdmX is actively involved in the degradation of both p53 and Hdm2.
Figures
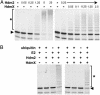
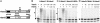



References
-
- Sharp, D. A., Kratowicz, S. A., Sank, M. J. & George, D. L. (1999) J. Biol. Chem. 274, 38189–38196. - PubMed
-
- Michael, D. & Oren, M. (2002) Curr. Opin. Genet. Dev. 12, 53–59. - PubMed
-
- Montes de Oca Luna, R., Wagner, D. S. & Lozano, G. (1995) Nature 378, 203–206. - PubMed
-
- Parant, J., Chavez-Reyes, A., Little, N. A., Yan, W., Reinke, V., Jochemsen, A. G. & Lozano, G. (2001) Nat. Genet. 29, 92–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

