ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response
- PMID: 14507999
- PMCID: PMC197305
- DOI: 10.1105/tpc.015412
ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response
Abstract
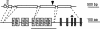
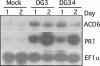
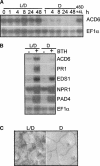
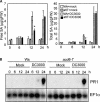
The previously reported Arabidopsis dominant gain-of-function mutant accelerated cell death6-1 (acd6-1) shows spontaneous cell death and increased disease resistance. acd6-1 also confers increased responsiveness to the major defense signal salicylic acid (SA). To further explore the role of ACD6 in the defense response, we cloned and characterized the gene. ACD6 encodes a novel protein with putative ankyrin and transmembrane regions. It is a member of one of the largest uncharacterized gene families in higher plants. Steady state basal expression of ACD6 mRNA required light, SA, and an intact SA signaling pathway. Additionally, ACD6 mRNA levels were increased in the systemic, uninfected tissue of Pseudomonas syringae-infected plants as well as in plants treated with the SA agonist benzothiazole (BTH). A newly isolated ACD6 loss-of-function mutant was less responsive to BTH and upon P. syringae infection had reduced SA levels and increased susceptibility. Conversely, plants overexpressing ACD6 showed modestly increased SA levels, increased resistance to P. syringae, and BTH-inducible and/or a low level of spontaneous cell death. Thus, ACD6 is a necessary and dose-dependent activator of the defense response against virulent bacteria and can activate SA-dependent cell death.
Figures




References
-
- Adam, L., and Somerville, S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9, 341–356. - PubMed
-
- Alvarez, M.E., Pennell, R.I., Meijer, P.J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. - PubMed
-
- Aviv, D.H., Rusterucci, C., Iii, B.F., Dietrich, R.A., Parker, J.E., and Dangl, J.L. (2002). Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 29, 381–391. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

