Lesions in phycoerythrin chromophore biosynthesis in Fremyella diplosiphon reveal coordinated light regulation of apoprotein and pigment biosynthetic enzyme gene expression
- PMID: 14508001
- PMCID: PMC197308
- DOI: 10.1105/tpc.015016
Lesions in phycoerythrin chromophore biosynthesis in Fremyella diplosiphon reveal coordinated light regulation of apoprotein and pigment biosynthetic enzyme gene expression
Abstract
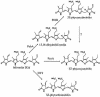
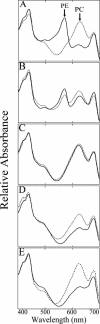
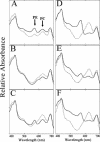
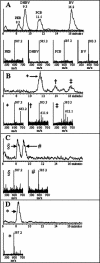
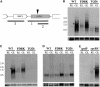
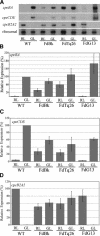
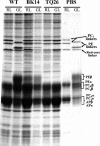
We have characterized the regulation of the expression of the pebAB operon, which encodes the enzymes required for phycoerythrobilin synthesis in the filamentous cyanobacterium Fremyella diplosiphon. The expression of the pebAB operon was found to be regulated during complementary chromatic adaptation, the system that controls the light responsiveness of genes that encode several light-harvesting proteins in F. diplosiphon. Our analyses of pebA mutants demonstrated that although the levels of phycoerythrin and its associated linker proteins decreased in the absence of phycoerythrobilin, there was no significant modulation of the expression of pebAB and the genes that encode phycoerythrin. Instead, regulation of the expression of these genes is coordinated at the level of RNA accumulation by the recently discovered activator CpeR.
Figures


References
-
- Anderson, L.K., and Toole, C.M. (1998). A model for early events in the assembly pathway of cyanobacterial phycobilisomes. Mol. Microbiol. 30, 467–474. - PubMed
-
- Arciero, D.M., Bryant, D.A., and Glazer, A.N. (1988). In vitro attachment of bilins to apophycocyanin. I. Specific covalent adduct formation at cysteinyl residues involved in phycocyanobilin binding in C-phycocyanin. J. Biol. Chem. 263, 18343–18349. - PubMed
-
- Beale, S.I., and Cornejo, J. (1991. a). Biosynthesis of phycobilisomes: 3(Z)-Phycoerythrobilin and 3(Z)-phycocyanobilin are intermediates in the formation of 3(E)-phycocyanobilin from biliverdin IXα. J. Biol. Chem. 266, 22333–22340. - PubMed
-
- Beale, S.I., and Cornejo, J. (1991. b). Biosynthesis of phycobilisomes: 15,16-Dihydrobiliverdin IXα is a partially reduced intermediate in the formation of phycobilins from biliverdin IXα. J. Biol. Chem. 266, 22341–22345. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

