Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction
- PMID: 14508006
- PMCID: PMC197304
- DOI: 10.1105/tpc.014498
Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction
Abstract
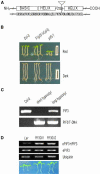
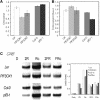
Phytochromes regulate various light responses through their interactions with different signaling proteins, such as phytochrome interacting factor 3 (PIF3). However, the physiological functions of PIF3 in light signaling are not yet fully understood. To increase our understanding of these roles, we characterized a T-DNA insertional pif3 mutant and transgenic plants overexpressing the full-length PIF3. Transgenic overexpressing lines displayed longer hypocotyls and smaller cotyledons under red light and reduced cotyledon opening under both red and far-red light, whereas the pif3 mutant showed the opposite phenotypes. The accumulation of anthocyanin and chlorophyll further indicated complicated features of PIF3 function. The accumulation of anthocyanin was increased and the content of chlorophyll was decreased in the overexpression lines. Our data indicate that PIF3 plays complex roles depending on the type of light response and the light conditions.
Figures

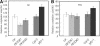


References
-
- Brownlie, P., Ceska, T., Lamers, M., Romier, C., Stier, G., Teo, H., and Suck, D. (1997). The crystal structure of an intact human Max-DNA complex: New insights into mechanisms of transcriptional control. Structure 5, 509–520. - PubMed
-
- Choe, S., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K.A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26, 573–582. - PubMed
-
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. - PubMed
-
- Ellenberger, T., Fass, D., Arnaud, M., and Harrison, S.C. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8, 970–980. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

