Microarray profiling of antibody responses against simian-human immunodeficiency virus: postchallenge convergence of reactivities independent of host histocompatibility type and vaccine regimen
- PMID: 14512560
- PMCID: PMC224970
- DOI: 10.1128/jvi.77.20.11125-11138.2003
Microarray profiling of antibody responses against simian-human immunodeficiency virus: postchallenge convergence of reactivities independent of host histocompatibility type and vaccine regimen
Abstract
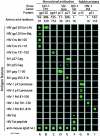
We developed antigen microarrays to profile the breadth, strength, and kinetics of epitope-specific antiviral antibody responses in vaccine trials with a simian-human immunodeficiency virus (SHIV) model for human immunodeficiency virus (HIV) infection. These arrays contained 430 distinct proteins and overlapping peptides spanning the SHIV proteome. In macaques vaccinated with three different DNA and/or recombinant modified vaccinia virus Ankara (rMVA) vaccines encoding Gag-Pol or Gag-Pol-Env, these arrays distinguished vaccinated from challenged macaques, identified three novel viral epitopes, and predicted survival. Following viral challenge, anti-SHIV antibody responses ultimately converged to target eight immunodominant B-cell regions in Env regardless of vaccine regimen, host histocompatibility type, and divergent T-cell specificities. After challenge, responses to nonimmunodominant epitopes were transient, while responses to dominant epitopes were gained. These data suggest that the functional diversity of anti-SHIV B-cell responses is highly limited in the presence of persisting antigen.
Figures






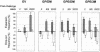

References
-
- Abacioglu, Y. H., T. R. Fouts, J. D. Laman, E. Claassen, S. H. Pincus, J. P. Moore, C. A. Roby, R. Kamin-Lewis, and G. K. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10:371-381. - PubMed
-
- Ahmad, A., and J. Menezes. 1996. Antibody-dependent cellular cytotoxicity in HIV infections. FASEB J. 10:258-266. - PubMed
-
- Alt, F. W., T. K. Blackwell, and G. D. Yancopoulos. 1987. Development of the primary antibody repertoire. Science 238:1079-1087. - PubMed
-
- Amara, R. R., and H. L. Robinson. 2002. A new generation of HIV vaccines. Trends Mol. Med. 8:489-495. - PubMed
-
- Amara, R. R., J. M. Smith, S. I. Staprans, D. C. Montefiori, F. Villinger, J. D. Altman, S. P. O'Neil, N. L. Kozyr, Y. Xu, L. S. Wyatt, P. L. Earl, J. G. Herndon, J. M. McNicholl, H. M. McClure, B. Moss, and H. L. Robinson. 2002. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J. Virol. 76:6138-6146. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

