Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate
- PMID: 14512572
- PMCID: PMC224963
- DOI: 10.1128/jvi.77.20.11244-11259.2003
Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate
Abstract
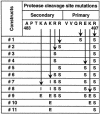
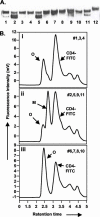
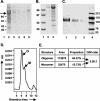
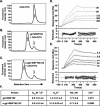
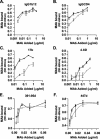
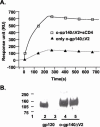
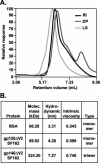
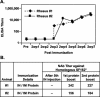
The envelope (Env) glycoprotein of human immunodeficiency virus type 1 (HIV-1) is the major target of neutralizing antibody responses and is likely to be a critical component of an effective vaccine against AIDS. Although monomeric HIV envelope subunit vaccines (gp120) have induced high-titer antibody responses and neutralizing antibodies against laboratory-adapted HIV-1 strains, they have failed to induce neutralizing antibodies against diverse heterologous primary HIV isolates. Most probably, the reason for this failure is that the antigenic structure(s) of these previously used immunogens does not mimic that of the functional HIV envelope, which is a trimer, and thus these immunogens do not elicit high titers of relevant functional antibodies. We recently reported that an Env glycoprotein immunogen (o-gp140SF162DeltaV2) containing a partial deletion in the second variable loop (V2) derived from the R5-tropic HIV-1 isolate SF162, when used in a DNA priming-protein boosting vaccine regimen in rhesus macaques, induced neutralizing antibodies against heterologous subtype B primary isolates as well as protection to the vaccinated animals upon challenge with pathogenic SHIV(SF162P4) virus. Here we describe the purification of this protein to homogeneity, its characterization as trimer, and its ability to induce primary isolate-neutralizing responses in rhesus macaques. Optimal mutations in the primary and secondary protease cleavage sites of the env gene were identified that resulted in the stable secretion of a trimeric Env glycoprotein in mammalian cell cultures. We determined the molecular mass and hydrodynamic radius (R(h)) using a triple detector analysis (TDA) system. The molecular mass of the oligomer was found to be 324 kDa, close to the expected M(w) of a HIV envelope trimer protein (330 kDa), and the hydrodynamic radius was 7.27 nm. Negative staining electron microscopy of o-gp140SF162DeltaV2 showed that it is a trimer with considerable structural flexibility and supported the data obtained by TDA. The structural integrity of the purified trimeric protein was also confirmed by determinations of its ability to bind the HIV receptor, CD4, and its ability to bind a panel of well-characterized neutralizing monoclonal antibodies. No deleterious effect of V2 loop deletion was observed on the structure and conformation of the protein, and several critical neutralization epitopes were preserved and well exposed on the purified o-gp140SF162DeltaV2 protein. In an intranasal priming and intramuscular boosting regimen, this protein induced high titers of functional antibodies, which neutralized the vaccine strain, i.e., SF162. These results highlight a potential role for the trimeric o-gp140SF162DeltaV2 Env immunogen in a successful HIV vaccine.
Figures



References
-
- Back, N. K., L. Smit, J. J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. - PubMed
-
- Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. - PMC - PubMed
-
- Barnett, S. W., S. Rajasekar, H. Legg, B. Doe, D. H. Fuller, J. R. Haynes, C. M. Walker, and K. S. Steimer. 1997. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine 15:869-873. - PubMed
-
- Barr, P. J., K. S. Steimer, E. A. Sabin, D. Parkes, C. George-Nascimento, J. C. Stephans, M. A. Powers, A. Gyenes, G. A. Van Nest, E. T. Miller, et al. 1987. Antigenicity and immunogenicity of domains of the human immunodeficiency virus (HIV) envelope polypeptide expressed in the yeast Saccharomyces cerevisiae. Vaccine 5:90-101. - PubMed
-
- Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

