Haplotypes in the dystrophin DNA segment point to a mosaic origin of modern human diversity
- PMID: 14513410
- PMCID: PMC1180505
- DOI: 10.1086/378777
Haplotypes in the dystrophin DNA segment point to a mosaic origin of modern human diversity
Abstract
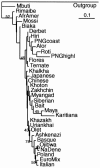
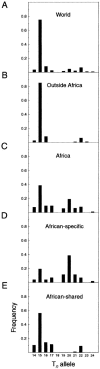
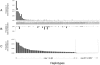
Although Africa has played a central role in human evolutionary history, certain studies have suggested that not all contemporary human genetic diversity is of recent African origin. We investigated 35 simple polymorphic sites and one T(n) microsatellite in an 8-kb segment of the dystrophin gene. We found 86 haplotypes in 1,343 chromosomes from around the world. Although a classical out-of-Africa topology was observed in trees based on the variant frequencies, the tree of haplotype sequences reveals three lineages accounting for present-day diversity. The proportion of new recombinants and the diversity of the T(n) microsatellite were used to estimate the age of haplotype lineages and the time of colonization events. The lineage that underwent the great expansion originated in Africa prior to the Upper Paleolithic (27,000-56,000 years ago). A second group, of structurally distinct haplotypes that occupy a central position on the tree, has never left Africa. The third lineage is represented by the haplotype that lies closest to the root, is virtually absent in Africa, and appears older than the recent out-of-Africa expansion. We propose that this lineage could have left Africa before the expansion (as early as 160,000 years ago) and admixed, outside of Africa, with the expanding lineage. Contemporary human diversity, although dominated by the recently expanded African lineage, thus represents a mosaic of different contributions.
Figures


Similar articles
-
Lactase haplotype diversity in the Old World.Am J Hum Genet. 2001 Jan;68(1):160-172. doi: 10.1086/316924. Epub 2000 Nov 28. Am J Hum Genet. 2001. PMID: 11095994 Free PMC article.
-
Haplotype trees and modern human origins.Am J Phys Anthropol. 2005;Suppl 41:33-59. doi: 10.1002/ajpa.20351. Am J Phys Anthropol. 2005. PMID: 16369961 Review.
-
Tracing genetic history of modern humans using X-chromosome lineages.Hum Genet. 2007 Dec;122(5):431-43. doi: 10.1007/s00439-007-0413-4. Epub 2007 Aug 7. Hum Genet. 2007. PMID: 17680273
-
Short tandem-repeat polymorphism/alu haplotype variation at the PLAT locus: implications for modern human origins.Am J Hum Genet. 2000 Oct;67(4):901-25. doi: 10.1086/303068. Epub 2000 Sep 13. Am J Hum Genet. 2000. PMID: 10986042 Free PMC article.
-
The peopling of the African continent and the diaspora into the new world.Curr Opin Genet Dev. 2014 Dec;29:120-32. doi: 10.1016/j.gde.2014.09.003. Curr Opin Genet Dev. 2014. PMID: 25461616 Free PMC article. Review.
Cited by
-
Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate.Proc Natl Acad Sci U S A. 2006 May 9;103(19):7234-9. doi: 10.1073/pnas.0508716103. Epub 2006 Apr 28. Proc Natl Acad Sci U S A. 2006. PMID: 16648268 Free PMC article.
-
Female-to-male breeding ratio in modern humans-an analysis based on historical recombinations.Am J Hum Genet. 2010 Mar 12;86(3):353-63. doi: 10.1016/j.ajhg.2010.01.029. Epub 2010 Feb 25. Am J Hum Genet. 2010. PMID: 20188344 Free PMC article.
-
Signatures of human European Palaeolithic expansion shown by resequencing of non-recombining X-chromosome segments.Eur J Hum Genet. 2017 Apr;25(4):485-492. doi: 10.1038/ejhg.2016.207. Epub 2017 Jan 25. Eur J Hum Genet. 2017. PMID: 28120839 Free PMC article.
-
Evidence for archaic adaptive introgression in humans.Nat Rev Genet. 2015 Jun;16(6):359-71. doi: 10.1038/nrg3936. Epub 2015 May 12. Nat Rev Genet. 2015. PMID: 25963373 Free PMC article. Review.
-
Current genetic methodologies in the identification of disaster victims and in forensic analysis.J Appl Genet. 2012 Feb;53(1):41-60. doi: 10.1007/s13353-011-0068-7. Epub 2011 Oct 15. J Appl Genet. 2012. PMID: 22002120 Free PMC article. Review.
References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the human dystrophin gene [accession number U94396])
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

