Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
- PMID: 14517232
- PMCID: PMC204477
- DOI: 10.1093/emboj/cdg494
Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Abstract
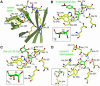
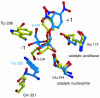
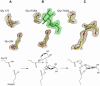
High-resolution crystal structures of alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T-6, a family 51 glycosidase, are described. The enzyme is a hexamer, and each monomer is organized into two domains: a (beta/alpha)8-barrel and a 12-stranded beta sandwich with jelly-roll topology. The structures of the Michaelis complexes with natural and synthetic substrates, and of the transient covalent arabinofuranosyl-enzyme intermediate represent two stable states in the double displacement mechanism, and allow thorough examination of the catalytic mechanism. The arabinofuranose sugar is tightly bound and distorted by an extensive network of hydrogen bonds. The two catalytic residues are 4.7 A apart, and together with other conserved residues contribute to the stabilization of the oxocarbenium ion-like transition state via charge delocalization and specific protein-substrate interactions. The enzyme is an anti-protonator, and a 1.7 A electrophilic migration of the anomeric carbon takes place during the hydrolysis.
Figures

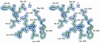
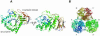

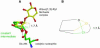
References
-
- Bourne Y. and Henrissat,B. (2001) Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol., 11, 593–600. - PubMed
-
- Collins P. and Ferrier,R. (1995) Monosaccharides, their Chemistry and their Roles in Natural Products. John Wiley and Sons Ltd, Chichester, UK.
-
- Davies G., Sinnott,M.L. and Withers,S.G. (1998a) Glycosyl Transfer. In Sinnott,M.L. (ed.), Comprehensive Biological Catalysis. Academic Press Limited, London, UK, Vol. 1, pp. 119–209.
-
- Davies G.J., Mackenzie,L., Varrot,A., Dauter,M., Brzozowski,A.M., Schulein,M. and Withers,S.G. (1998b) Snapshots along an enzymatic reaction coordinate: analysis of a retaining β-glycoside hydrolase. Biochemistry, 37, 11707–11713. - PubMed
-
- Debeche T., Bliard,C., Debeire,P. and O’Donohue,M.J. (2002) Probing the catalytically essential residues of the α-l-arabinofuranosidase from Thermobacillus xylanilyticus. Protein Eng., 15, 21–28. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

