Single-molecule imaging of cooperative assembly of gamma-hemolysin on erythrocyte membranes
- PMID: 14517236
- PMCID: PMC204481
- DOI: 10.1093/emboj/cdg498
Single-molecule imaging of cooperative assembly of gamma-hemolysin on erythrocyte membranes
Abstract
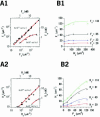
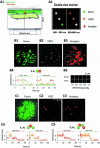
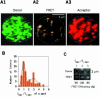
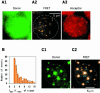
Single-molecule fluorescence imaging was used to investigate assembly of Staphylococcus aureus LukF and HS monomers into pore-forming oligomers (gamma-hemolysin) on erythrocyte membranes. We distinguished the hetero-oligomers from the monomers, as indicated by fluorescence resonance energy transfer between different dyes attached to monomeric subunits. The stoichiometry of LukF (donor) and HS (acceptor) subunits in oligomers was deduced from the acceptor emission intensities during energy transfer and by direct acceptor excitation, respectively. Based on populations of monomeric and oligomeric intermediates, we estimated 11 sequential equilibrium constants for the assembly pathway, beginning with membrane binding of monomers, proceeding through single pore oligomerization, and culminating in the formation of clusters of pores. Several stages are highly cooperative, critically enhancing the efficiency of assembly.
Figures


References
-
- Alberts B., Alexander,J., Lewis,J., Raff,M., Roberts,K. and Walter,P. (2002) Protein. In Albert,B. (ed.), Molecular Biology of the Cell, 4th edn. Garland Science, New York, NY, pp. 147–162.
-
- Alberts B., Alexander,J., Lewis,J., Raff,M., Roberts,K. and Walter,P. (2002) The cytoskelton. In Albert,B. (ed.), Molecular Biology of the Cell, 4th edn. Garland Science, New York, NY, pp. 907–915.
-
- Comai M. et al. (2002) Protein engineering modulates the transport properties and ion selectivity of the pores formed by staphylococcal γ-hemolysins in lipid membranes. Mol. Microbiol., 44, 1251–1267. - PubMed
-
- Daugherty M.A., Brenowitz,M. and Fried,M.G. (1999) The TATA-binding protein from Saccharomyces cerevisiae oligomerizes in solution at micromolar concentrations to form tetramers and octamers. J. Mol. Biol., 285, 1389–1399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

