SseG, a virulence protein that targets Salmonella to the Golgi network
- PMID: 14517239
- PMCID: PMC204495
- DOI: 10.1093/emboj/cdg517
SseG, a virulence protein that targets Salmonella to the Golgi network
Abstract
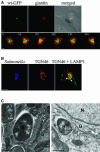
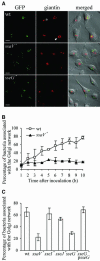
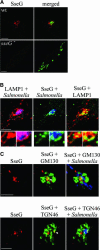
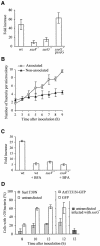
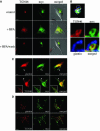
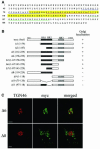
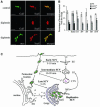
Intracellular replication of the bacterial pathogen Salmonella enterica occurs in membrane-bound compartments called Salmonella-containing vacuoles (SCVs). Maturation of the SCV has been shown to occur by selective interactions with the endocytic pathway. We show here that after invasion of epithelial cells and migration to a perinuclear location, the majority of SCVs become surrounded by membranes of the Golgi network. This process is dependent on the Salmonella pathogenicity island 2 type III secretion system effector SseG. In infected cells, SseG was associated with the SCV and peripheral punctate structures. Only bacterial cells closely associated with the Golgi network were able to multiply; furthermore, mutation of sseG or disruption of the Golgi network inhibited intracellular bacterial growth. When expressed in epithelial cells, SseG co-localized extensively with markers of the trans-Golgi network. We identify a Golgi-targeting domain within SseG, and other regions of the protein that are required for localization of bacteria to the Golgi network. Therefore, replication of Salmonella in epithelial cells is dependent on simultaneous and selective interactions with both endocytic and secretory pathways.
Figures
References
-
- Berger E.G., Aegerter,E., Mandel,T. and Hauri,H.P. (1986) Monoclonal antibodies to soluble, human milk galactosyltransferase (lactose synthase A protein). Carbohydr. Res., 149, 23–33. - PubMed
-
- Beuzón C.R., Salcedo,S.P. and Holden,D.W. (2002) Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology, 148, 2705–2715. - PubMed
-
- Brumell J.H., Tang,P., Mills,S.D. and Finlay,B.B. (2001) Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic, 2, 643–653. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

