Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival
- PMID: 14517290
- PMCID: PMC230321
- DOI: 10.1128/MCB.23.20.7198-7209.2003
Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival
Abstract
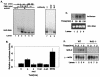
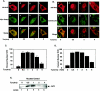
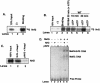
Activation of PERK following the accumulation of unfolded proteins in the endoplasmic reticulum (ER) promotes translation inhibition and cell cycle arrest. PERK function is essential for cell survival following exposure of cells to ER stress, but the mechanisms whereby PERK signaling promotes cell survival are not thoroughly understood. We have identified the Nrf2 transcription factor as a novel PERK substrate. In unstressed cells, Nrf2 is maintained in the cytoplasm via association with Keap1. PERK-dependent phosphorylation triggers dissociation of Nrf2/Keap1 complexes and inhibits reassociation of Nrf2/Keap1 complexes in vitro. Activation of PERK via agents that trigger the unfolded protein response is both necessary and sufficient for dissociation of cytoplasmic Nrf2/Keap1 and subsequent Nrf2 nuclear import. Finally, we demonstrate that cells harboring a targeted deletion of Nrf2 exhibit increased cell death relative to wild-type counterparts following exposure to ER stress. Our data demonstrate that Nrf2 is a critical effector of PERK-mediated cell survival.
Figures





References
-
- Andrews, N. C., H. Erdjument-Bromage, M. B. Davidson, P. Tempst, and S. H. Orkin. 1993. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362:722-728. - PubMed
-
- Berlanga, J. J., J. Santoyo, and C. Haro. 1999. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur. J. Biochem. 265:754-762. - PubMed
-
- Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
