The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides
- PMID: 14517307
- PMCID: PMC230319
- DOI: 10.1128/MCB.23.20.7403-7414.2003
The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides
Abstract
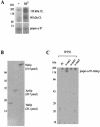
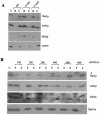
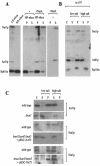
The majority of cytosolic proteins in eukaryotes contain a covalently linked acetyl moiety at their very N terminus. The mechanism by which the acetyl moiety is efficiently transferred to a large variety of nascent polypeptides is currently only poorly understood. Yeast N(alpha)-acetyltransferase NatA, consisting of the known subunits Nat1p and the catalytically active Ard1p, recognizes a wide range of sequences and is thought to act cotranslationally. We found that NatA was quantitatively bound to ribosomes via Nat1p and contained a previously unrecognized third subunit, the N(alpha)-acetyltransferase homologue Nat5p. Nat1p not only anchored Ard1p and Nat5p to the ribosome but also was in close proximity to nascent polypeptides, independent of whether they were substrates for N(alpha)-acetylation or not. Besides Nat1p, NAC (nascent polypeptide-associated complex) and the Hsp70 homologue Ssb1/2p interact with a variety of nascent polypeptides on the yeast ribosome. A direct comparison revealed that Nat1p required longer nascent polypeptides for interaction than NAC and Ssb1/2p. Delta nat1 or Delta ard1 deletion strains were temperature sensitive and showed derepression of silent mating type loci while Delta nat5 did not display any obvious phenotype. Temperature sensitivity and derepression of silent mating type loci caused by Delta nat1 or Delta ard1 were partially suppressed by overexpression of SSB1. The combination of data suggests that Nat1p presents the N termini of nascent polypeptides for acetylation and might serve additional roles during protein synthesis.
Figures




References
-
- Andrade, M. A., C. P. Ponting, T. J. Gibson, and P. Bork. 2000. Homology-based method for identification of protein repeats using statistical significance estimates. J. Mol. Biol. 298:521-537. - PubMed
-
- Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279-1287. - PubMed
-
- Arnold, R. J., B. Polevoda, J. P. Reilly, and F. Sherman. 1999. The action of N-terminal acetyltransferases on yeast ribosomal proteins. J. Biol. Chem. 274:37035-37040. - PubMed
-
- Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
