Identifying biological themes within lists of genes with EASE
- PMID: 14519205
- PMCID: PMC328459
- DOI: 10.1186/gb-2003-4-10-r70
Identifying biological themes within lists of genes with EASE
Abstract
EASE is a customizable software application for rapid biological interpretation of gene lists that result from the analysis of microarray, proteomics, SAGE and other high-throughput genomic data. The biological themes returned by EASE recapitulate manually determined themes in previously published gene lists and are robust to varying methods of normalization, intensity calculation and statistical selection of genes. EASE is a powerful tool for rapidly converting the results of functional genomics studies from 'genes' to 'themes'.
Figures
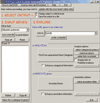
References
-
- Heller MJ. DNA microarray technology: devices, systems, and applications. Annu Rev Biomed Eng. 2002;4:129–153. - PubMed
-
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32 Suppl:496–501. - PubMed
-
- Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32 Suppl:502–508. - PubMed
-
- Pruitt KD, Katz KS, Sicotte H, Maglott DR. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

