Epidemic and nonepidemic multidrug-resistant Enterococcus faecium
- PMID: 14519248
- PMCID: PMC3016763
- DOI: 10.3201/eid0909.020383
Epidemic and nonepidemic multidrug-resistant Enterococcus faecium
Abstract
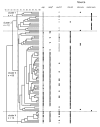
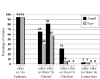
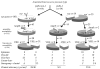
The epidemiology of vancomycin-resistant Entero- coccus faecium (VREF) in Europe is characterized by a large community reservoir. In contrast, nosocomial outbreaks and infections (without a community reservoir) characterize VREF in the United States. Previous studies demonstrated host-specific genogroups and a distinct genetic lineage of VREF associated with hospital outbreaks, characterized by the variant esp-gene and a specific allele-type of the purK housekeeping gene (purK1). We investigated the genetic relatedness of vanA VREF (n=108) and vancomycin-susceptible E. faecium (VSEF) (n=92) from different epidemiologic sources by genotyping, susceptibility testing for ampicillin, sequencing of purK1, and testing for presence of esp. Clusters of VSEF fit well into previously described VREF genogroups, and strong associations were found between VSEF and VREF isolates with resistance to ampicillin, presence of esp, and purK1. Genotypes characterized by presence of esp, purK1, and ampicillin resistance were most frequent among outbreak-associated isolates and almost absent among community surveillance isolates. Vancomycin-resistance was not specifically linked to genogroups. VREF and VSEF from different epidemiologic sources are genetically related; evidence exists for nosocomial selection of a subtype of E. faecium, which has acquired vancomycin-resistance through horizontal transfer.
Figures
References
-
- van der Auwera P, Pensart N, Korten V, Murray BE, Leclercq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis. 1996;173:1129–36. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
