KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans
- PMID: 14522947
- PMCID: PMC218079
- DOI: 10.1101/gad.1126303
KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans
Abstract
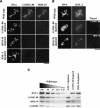
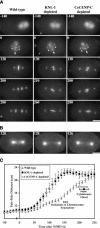
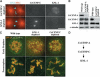
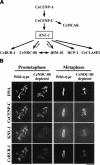
Segregation of the replicated genome during cell division requires kinetochores, mechanochemical organelles that assemble on mitotic chromosomes to connect them to spindle microtubules. CENP-A, a histone H3 variant, and CENP-C, a conserved structural protein, form the DNA-proximal foundation for kinetochore assembly. Using RNA interference-based genomics in Caenorhabditis elegans, we identified KNL-1, a novel kinetochore protein whose depletion, like that of CeCENP-A or CeCENP-C, leads to a "kinetochore-null" phenotype. KNL-1 is downstream of CeCENP-A and CeCENP-C in a linear assembly hierarchy. In embryonic extracts, KNL-1 exhibits substoichiometric interactions with CeCENP-C and forms a near-stoichiometric complex with CeNDC-80 and HIM-10, the C. elegans homologs of Ndc80p/HEC1p and Nuf2p-two widely conserved outer kinetochore components. However, CeNDC-80 and HIM-10 are not functionally equivalent to KNL-1 because their inhibition, although preventing formation of a mechanically stable kinetochore-microtubule interface and causing chromosome missegregation, does not result in a kinetochore-null phenotype. The greater functional importance of KNL-1 may be due to its requirement for targeting multiple components of the outer kinetochore, including CeNDC-80 and HIM-10. Thus, KNL-1 plays a central role in translating the initiation of kinetochore assembly by CeCENP-A and CeCENP-C into the formation of a functional microtubule-binding interface.
Figures



References
-
- Akhmanova A., Hoogenraad, C.C., Drabek, K., Stepanova, T., Dortland, B., Verkerk, T., Vermeulen, W., Burgering, B.M., De Zeeuw, C.I., Grosveld, F., et al. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104: 923-935. - PubMed
-
- Cheeseman I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates III, J.R., Chan, C.S., Drubin, D.G., and Barnes, G. 2002a. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163-172. - PubMed
-
- Cleveland D.W., Mao, Y., and Sullivan, K.F. 2003. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 112: 407-421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
