Sp8 is crucial for limb outgrowth and neuropore closure
- PMID: 14526104
- PMCID: PMC218735
- DOI: 10.1073/pnas.2134310100
Sp8 is crucial for limb outgrowth and neuropore closure
Abstract
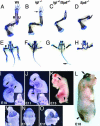
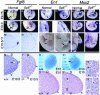
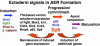
In this report we describe the developmental expression and function of Sp8, a member of the Sp family of zinc finger transcription factors, and provide evidence that the legless transgene insertional mutant is a hypomorphic allele of the Sp8 gene. Sp8 is expressed during embryogenesis in the forming apical ectodermal ridge (AER), restricted regions of the central nervous system, and tail bud. Targeted deletion of the Sp8 gene gives a striking phenotype, with severe truncation of both forelimbs and hindlimbs, absent tail, as well as defects in anterior and posterior neuropore closure leading to exencephaly and spina bifida. Outgrowth of the limb depends on formation of the AER, a signaling center that forms at the limb bud apex. In Sp8 mutants, the AER precursor cells are induced and initially express multiple appropriate marker genes, but expression of these genes is not maintained and progression to a mature AER is blocked. These observations indicate that Sp8 functions downstream of Wnt3, Fgf10, and Bmpr1a in the signaling cascade that mediates AER formation.
Figures

References
-
- Ahn, K., Mishina, Y., Hanks, M. C., Behringer, R. R. & Crenshaw, E. B. I. (2001) Development (Cambridge, U.K.) 128, 4449–4461. - PubMed
-
- Xu, X., Weinstein, M., Li, Cuiling, Naski, M., Cohen, R. I., Ornitz, D. M., Leder, P. & Deng, C. (1998) Development (Cambridge, U.K.) 125, 753–765. - PubMed
-
- Moerlooze, D. L., Spencer-Dene, B., Revest, J.-M., Hajihosseini, M., Rosewell, I. & Dickson, C. (2000) Development (Cambridge, U.K.) 127, 483–492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

