Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis
- PMID: 14526118
- PMCID: PMC281622
- DOI: 10.1104/pp.103.024182
Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis
Abstract
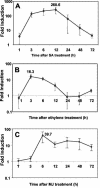
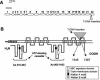
The ATP-binding cassette (ABC) transporters are encoded by large gene families in plants. Although these proteins are potentially involved in a number of diverse plant processes, currently, very little is known about their actual functions. In this paper, through a cDNA microarray screening of anonymous cDNA clones from a subtractive library, we identified an Arabidopsis gene (AtPDR12) putatively encoding a member of the pleiotropic drug resistance (PDR) subfamily of ABC transporters. AtPDR12 displayed distinct induction profiles after inoculation of plants with compatible and incompatible fungal pathogens and treatments with salicylic acid, ethylene, or methyl jasmonate. Analysis of AtPDR12 expression in a number of Arabidopsis defense signaling mutants further revealed that salicylic acid accumulation, NPR1 function, and sensitivity to jasmonates and ethylene were all required for pathogen-responsive expression of AtPDR12. Germination assays using seeds from an AtPDR12 insertion line in the presence of sclareol resulted in lower germination rates and much stronger inhibition of root elongation in the AtPDR12 insertion line than in wild-type plants. These results suggest that AtPDR12 may be functionally related to the previously identified ABC transporters SpTUR2 and NpABC1, which transport sclareol. Our data also point to a potential role for terpenoids in the Arabidopsis defensive armory.
Figures


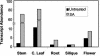




References
-
- Aubourg S, Lecharny A, Bohlman J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genome 267: 730–745 - PubMed
-
- Bailey JA, Vincent GG, Burden RS (1974) Diterpenes from Nicotiana glutinosa and their effect on fungal growth. J Gen Microbiol 85: 57–64 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

