Biomolecular hydration: from water dynamics to hydrodynamics
- PMID: 14528004
- PMCID: PMC218725
- DOI: 10.1073/pnas.2033320100
Biomolecular hydration: from water dynamics to hydrodynamics
Abstract
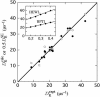
Thermally driven rotational and translational diffusion of proteins and other biomolecules is governed by frictional coupling to their solvent environment. Prediction of this coupling from biomolecular structures is a longstanding biophysical problem, which cannot be solved without knowledge of water dynamics in an interfacial region comparable to the dry protein in volume. Efficient algorithms have been developed for solving the hydrodynamic equations of motion for atomic-resolution biomolecular models, but experimental diffusion coefficients can be reproduced only by postulating hundreds of rigidly bound water molecules. This static picture of biomolecular hydration is fundamentally inconsistent with magnetic relaxation dispersion experiments and molecular dynamics simulations, which both reveal a highly dynamic interface where rotation and exchange of nearly all water molecules are several orders of magnitude faster than biomolecular diffusion. Here, we resolve this paradox by means of a dynamic hydration model that explicitly links protein hydrodynamics to hydration dynamics. With the aid of this model, bona fide structure-based predictions of global biomolecular dynamics become possible, as demonstrated here for a set of 16 proteins for which accurate experimental rotational diffusion coefficients are available.
Figures
References
-
- Zwanzig, R. (2001) Nonequilibrium Statistical Mechanics (Oxford Univ. Press, New York).
-
- Einstein, A. (1956) in Investigations on the Theory of Brownian Movement, ed. Fürth, R. (Dover, New York).
-
- Landau, L. D. & Lifshitz, E. M. (1959) Fluid Mechanics (Pergamon, Oxford).
-
- Perrin, F. (1936) J. Phys. Radium 7, 1–11.
-
- Buck, M., Boyd, J., Redfield, C., MacKenzie, D. A., Jeenes, D. J., Archer, D. B. & Dobson, C. M. (1995) Biochemistry 34, 4041–4055. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

