Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast
- PMID: 14528010
- PMCID: PMC307528
- DOI: 10.1091/mbc.e03-08-0586
Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast
Abstract
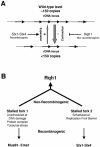
In most eukaryotes, genes encoding ribosomal RNAs (rDNA) are clustered in long tandem head-to-tail repeats. Studies of Saccharomyces cerevisiae have indicated that rDNA copy number is maintained through recombination events associated with site-specific blockage of replication forks (RFs). Here, we describe two Schizosaccharomyces pombe proteins, homologs of S. cerevisiae Slx1 and Slx4, as subunits of a novel type of endonuclease that maintains rDNA copy number. The Slx1-Slx4-dependent endonuclease introduces single-strand cuts in duplex DNA on the 3' side of junctions with single-strand DNA. Deletion of Slx1 or Rqh1 RecQ-like DNA helicase provokes rDNA contraction, whereas simultaneous elimination of Slx1-Slx4 endonuclease and Rqh1 is lethal. Slx1 associates with chromatin at two foci characteristic of the two rDNA repeat loci in S. pombe. We propose a model in which the Slx1-Slx4 complex is involved in the control of the expansion and contraction of the rDNA loci by initiating recombination events at stalled RFs.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

