Ligand crowding at a nascent signal sequence
- PMID: 14530384
- PMCID: PMC2173441
- DOI: 10.1083/jcb.200306069
Ligand crowding at a nascent signal sequence
Abstract
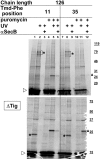
We have systematically analyzed the molecular environment of the signal sequence of a growing secretory protein from Escherichia coli using a stage- and site-specific cross-linking approach. Immediately after emerging from the ribosome, the signal sequence of pOmpA is accessible to Ffh, the protein component of the bacterial signal recognition particle, and to SecA, but it remains attached to the surface of the ribosome via protein L23. These contacts are lost upon further growth of the nascent chain, which brings the signal sequence into sole proximity to the chaperone Trigger factor (TF). In its absence, nascent pOmpA shows extended contacts with L23, and even long chains interact in these conditions proficiently with Ffh. Our results suggest that upon emergence from the ribosome, the signal sequence of an E. coli secretory protein gradually becomes sequestered by TF. Although TF thereby might control the accessibility of pOmpA's signal sequence to Ffh and SecA, it does not influence interaction of pOmpA with SecB.
Figures







Similar articles
-
Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor.Proc Natl Acad Sci U S A. 2004 May 25;101(21):7902-6. doi: 10.1073/pnas.0402231101. Epub 2004 May 17. Proc Natl Acad Sci U S A. 2004. PMID: 15148364 Free PMC article.
-
Selective SecA association with signal sequences in ribosome-bound nascent chains: a potential role for SecA in ribosome targeting to the bacterial membrane.J Biol Chem. 2005 Nov 11;280(45):37930-40. doi: 10.1074/jbc.M509100200. Epub 2005 Aug 23. J Biol Chem. 2005. PMID: 16120599
-
Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor.EMBO J. 2000 Jan 4;19(1):134-43. doi: 10.1093/emboj/19.1.134. EMBO J. 2000. PMID: 10619852 Free PMC article.
-
Recognition of ligands by SecB, a molecular chaperone involved in bacterial protein export.Philos Trans R Soc Lond B Biol Sci. 1993 Mar 29;339(1289):343-52; discussion 352-4. doi: 10.1098/rstb.1993.0033. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8098539 Review.
-
The signal recognition particle and its interactions during protein targeting.Curr Opin Struct Biol. 2005 Feb;15(1):116-25. doi: 10.1016/j.sbi.2005.01.013. Curr Opin Struct Biol. 2005. PMID: 15718142 Review.
Cited by
-
The Dynamic SecYEG Translocon.Front Mol Biosci. 2021 Apr 15;8:664241. doi: 10.3389/fmolb.2021.664241. eCollection 2021. Front Mol Biosci. 2021. PMID: 33937339 Free PMC article. Review.
-
TatB functions as an oligomeric binding site for folded Tat precursor proteins.Mol Biol Cell. 2010 Dec;21(23):4151-61. doi: 10.1091/mbc.E10-07-0585. Epub 2010 Oct 6. Mol Biol Cell. 2010. PMID: 20926683 Free PMC article.
-
Delivering proteins for export from the cytosol.Nat Rev Mol Cell Biol. 2009 Apr;10(4):255-64. doi: 10.1038/nrm2657. Nat Rev Mol Cell Biol. 2009. PMID: 19305415 Review.
-
Signal recognition particle and SecA cooperate during export of secretory proteins with highly hydrophobic signal sequences.PLoS One. 2014 Apr 9;9(4):e92994. doi: 10.1371/journal.pone.0092994. eCollection 2014. PLoS One. 2014. PMID: 24717922 Free PMC article.
-
How Quality Control Systems AID Sec-Dependent Protein Translocation.Front Mol Biosci. 2021 Apr 13;8:669376. doi: 10.3389/fmolb.2021.669376. eCollection 2021. Front Mol Biosci. 2021. PMID: 33928127 Free PMC article. Review.
References
-
- Beha, D., S. Deitermann, M. Muller, and H.G. Koch. 2003. Export of β-lactamase is independent of the signal recognition particle. J. Biol. Chem. 278:22161–22167. - PubMed
-
- Behrmann, M., H.G. Koch, T. Hengelage, B. Wieseler, H.K. Hoffschulte, and M. Muller. 1998. Requirements for the translocation of elongation-arrested, ribosome-associated OmpA across the plasma membrane of Escherichia coli. J. Biol. Chem. 273:13898–13904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

