The energetic cost of domain reorientation in maltose-binding protein as studied by NMR and fluorescence spectroscopy
- PMID: 14530390
- PMCID: PMC240681
- DOI: 10.1073/pnas.2134311100
The energetic cost of domain reorientation in maltose-binding protein as studied by NMR and fluorescence spectroscopy
Abstract
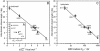
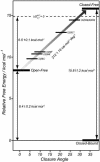
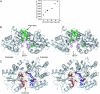
Maltose-binding protein (MBP) is a two-domain protein that undergoes a ligand-mediated conformational rearrangement from an "open" to a "closed" structure on binding to maltooligosaccharides. To characterize the energy landscape associated with this transition, we have generated five variants of MBP with mutations located in the hinge region of the molecule. Residual dipolar couplings, measured in the presence of a weak alignment medium, have been used to establish that the average structures of the mutant proteins are related to each other by domain rotation about an invariant axis, with the rotation angle varying from 5 degrees to 28 degrees. Additionally, the domain orientations observed in the wild-type apo and ligand-bound (maltose, maltotriose, etc.) structures are related through a rotation of 35 degrees about the same axis. Remarkably, the free energy of unfolding, measured by equilibrium denaturation experiments and monitored by fluorescence spectroscopy, shows a linear correlation with the rotation angle, with the stability of the (apo)protein decreasing with domain closure by 212 +/- 16 cal mol-1 per degree of rotation. The apparent binding energy for maltose also shows a similar correlation with the interdomain angle, suggesting that the mutations, as they relate to binding, affect predominantly the ligand-free structure. The linearity of the energy change is interpreted in terms of an increase in the extent of hydrophobic surface that becomes solvent accessible on closure. The combination of structural, stability, and binding data allows separation of the energetics of domain reorientation from ligand binding. This work presents a near quantitative structure-energy-binding relationship for a series of mutants of MBP, illustrating the power of combined studies involving protein engineering and solution NMR spectroscopy.
Figures

Comment in
-
The energetics of structural change in maltose-binding protein.Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12529-30. doi: 10.1073/pnas.2335923100. Epub 2003 Oct 20. Proc Natl Acad Sci U S A. 2003. PMID: 14569013 Free PMC article. No abstract available.
References
-
- Gerstein, M., Lesk, A. M. & Chothia, C. (1994) Biochemistry 33 6739–6749. - PubMed
-
- Lawson, C. L., Zhang, R., Schevitz, R. W., Otwinowski, Z., Joachimiak, A. & Sigler, P. B. (1988) Proteins Struct. Funct. Genet. 3 18–31. - PubMed
-
- Ikura, M., Clore, G. M., Gronenborn, A. M., Zhu, G., Klee, C. B. & Bax, A. (1992) Science 256 632–638. - PubMed
-
- Oh, B. H., Pandit, J., Kang, C. H., Nikaido, K., Gokcen, S., Ames, G. F.-L. & Kim, S. H. (1993) J. Biol. Chem. 268 11348–11355. - PubMed
-
- Stillman, T. J., Baker, B. J., Britton, K. L. & Rice, D. W. (1993) J. Mol. Biol. 234 1131–1139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

