Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays
- PMID: 14530437
- PMCID: PMC219483
- DOI: 10.1093/nar/gkg806
Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays
Abstract
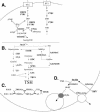
In the protozoan parasite Leishmania, drug resistance can be a complex phenomenon. Several metabolic pathways and membrane transporters are implicated in the resistance phenotype. To monitor the expression of these genes, we generated custom DNA microarrays with PCR fragments corresponding to 44 genes involved with drug resistance. Transcript profiling of arsenite and antimony resistant mutants with these arrays pinpointed a number of genes overexpressed in mutants, including the ABC transporter PGPA, the glutathione biosynthesis genes gamma-glutamylcysteine synthetase (GSH1) and the glutathione synthetase (GSH2). Competitive hybridisations with total RNA derived from sensitive and methotrexate resistant cells revealed the overexpression of genes coding for dihydrofolate reductase (DHFR-TS), pteridine reductase (PTR1) and S-adenosylmethionine synthase (MAT2) and a down regulation of one gene of the folate transporter (FT) family. By labelling the DNA of sensitive and resistant parasites we could also detect several gene amplification events using DNA microarrays including the amplification of the S-adenosyl homocysteine hydrolase gene (SAHH). Alteration in gene expression detected by microarrays was validated by northern blot analysis, while Southern blots indicated that most genes overexpressed were also amplified, although other mechanisms were also present. The microarrays were useful in the study of resistant parasites to pinpoint several genes linked to drug resistance.
Figures


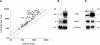

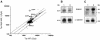
References
-
- Herwaldt B.L. (1999) Leishmaniasis. Lancet, 354, 1191–1199. - PubMed
-
- Guerin P.J., Olliaro,P., Sundar,S., Boelaert,M., Croft,S.L., Desjeux,P., Wasunna,M.K. and Bryceson,A.D. (2002) Visceral leishmaniasis: current status of control, diagnosis and treatment and a proposed research and development agenda. Lancet Infect. Dis., 2, 494–501. - PubMed
-
- Sundar S., More,D.K., Singh,M.K., Singh,V.P., Sharma,S., Makharia,A., Kumar,P.C. and Murray,H.W. (2000) Failure of pentavalent antimony in visceral leishmaniasis in India: report from the Center of the Indian Epidemic. Clin. Infect. Dis., 31, 1104–1107. - PubMed
-
- Ouellette M. and Ward,S. (2002) Drug resistance in parasites. In Marr,J., Nielsen,T. and Komuniecki,R. (eds), Molecular Medical Parasitology. Academic Press, San Diego, pp. 395–430.
-
- Shaked-Mishan P., Ulrich,N., Ephros,M. and Zilberstein,D. (2001) Novel intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J. Biol. Chem., 276, 3971–3976. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

