Sequence-specific fluorescent labeling of double-stranded DNA observed at the single molecule level
- PMID: 14530458
- PMCID: PMC219493
- DOI: 10.1093/nar/gng125
Sequence-specific fluorescent labeling of double-stranded DNA observed at the single molecule level
Abstract
Fluorescent labeling of a short sequence of double-stranded DNA (dsDNA) was achieved by ligating a labeled dsDNA fragment to a stem-loop triplex forming oligonucleotide (TFO). After the TFO has wound around the target sequence by ligand-induced triple helix formation, its extremities hybridize to each other, leaving a dangling single-stranded sequence, which is then ligated to a fluorescent dsDNA fragment using T4 DNA ligase. A non-repeated 15 bp sequence present on lambda DNA was labeled and visualized by fluorescence microscopy after DNA combing. The label was found to be attached at a specific position located at 4.2 +/- 0.5 kb from one end of the molecule, in agreement with the location of the target sequence for triple helix formation (4.4 kb from one end). In addition, an alternative combing process was noticed in which a DNA molecule becomes attached to the combing slide from the label rather than from one of its ends. The method described herein provides a new tool for the detection of very short sequences of dsDNA and offers various perspectives in the micromanipulation of single DNA molecules.
Figures
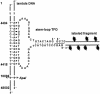


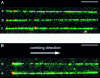

References
-
- Tiner W.J. Sr, Potaman,V.N., Sinden,R.R. and Lyubchenko,Y.L. (2001) The structure of intramolecular triplex DNA: atomic force microscopy study. J. Mol. Biol., 314, 353–357. - PubMed
-
- Strick T., Allemand,J., Croquette,V. and Bensimon,D. (2000) Twisting and stretching single DNA molecules. Prog. Biophys. Mol. Biol., 74, 115–140. - PubMed
-
- Wang M.D., Schnitzer,M.J., Yin,H., Landick,R., Gelles,J. and Block,S.M. (1998) Force and velocity measured for single molecules of RNA polymerase. Science, 282, 902–907. - PubMed
-
- Strick T.R., Croquette,V. and Bensimon,D. (2000) Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature, 404, 901–904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

