Diversity of nitrile hydratase and amidase enzyme genes in Rhodococcus erythropolis recovered from geographically distinct habitats
- PMID: 14532022
- PMCID: PMC201182
- DOI: 10.1128/AEM.69.10.5754-5766.2003
Diversity of nitrile hydratase and amidase enzyme genes in Rhodococcus erythropolis recovered from geographically distinct habitats
Abstract
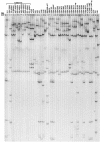
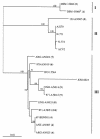
A molecular screening approach was developed in order to amplify the genomic region that codes for the alpha- and beta-subunits of the nitrile hydratase (NHase) enzyme in rhodococci. Specific PCR primers were designed for the NHase genes from a collection of nitrile-degrading actinomycetes, but amplification was successful only with strains identified as Rhodococcus erythropolis. A hydratase PCR product was also obtained from R. erythropolis DSM 43066(T), which did not grow on nitriles. Southern hybridization of other members of the nitrile-degrading bacterial collection resulted in no positive signals other than those for the R. erythropolis strains used as positive controls. PCR-restriction fragment length polymorphism-single-strand conformational polymorphism (PRS) analysis of the hydratases in the R. erythropolis strains revealed unique patterns that mostly correlated with distinct geographical sites of origin. Representative NHases were sequenced, and they exhibited more than 92.4% similarity to previously described NHases. The phylogenetic analysis and deduced amino acid sequences suggested that the novel R. erythropolis enzymes belonged to the iron-type NHase family. Some different residues in the translated sequences were located near the residues involved in the stabilization of the NHase active site, suggesting that the substitutions could be responsible for the different enzyme activities and substrate specificities observed previously in this group of actinomycetes. A similar molecular screening analysis of the amidase gene was performed, and a correlation between the PRS patterns and the geographical origins identical to the correlation found for the NHase gene was obtained, suggesting that there was coevolution of the two enzymes in R. erythropolis. Our findings indicate that the NHase and amidase genes present in geographically distinct R. erythropolis strains are not globally mixed.
Figures


Similar articles
-
Characterization of nitrile hydratase genes cloned by DNA screening from Rhodococcus erythropolis.Biosci Biotechnol Biochem. 1993 Aug;57(8):1323-8. doi: 10.1271/bbb.57.1323. Biosci Biotechnol Biochem. 1993. PMID: 7764017
-
Expression control of nitrile hydratase and amidase genes in Rhodococcus erythropolis and substrate specificities of the enzymes.Antonie Van Leeuwenhoek. 2014 Jun;105(6):1179-90. doi: 10.1007/s10482-014-0179-3. Epub 2014 Apr 30. Antonie Van Leeuwenhoek. 2014. PMID: 24781748
-
Searching for nitrile hydratase using the Consensus-Degenerate Hybrid Oligonucleotide Primers strategy.J Basic Microbiol. 2004;44(3):203-14. doi: 10.1002/jobm.200310340. J Basic Microbiol. 2004. PMID: 15162394
-
Fe-type nitrile hydratase.J Inorg Biochem. 2001 Feb;83(4):247-53. doi: 10.1016/s0162-0134(00)00171-9. J Inorg Biochem. 2001. PMID: 11293544 Review.
-
Metalloenzyme nitrile hydratase: structure, regulation, and application to biotechnology.Nat Biotechnol. 1998 Aug;16(8):733-6. doi: 10.1038/nbt0898-733. Nat Biotechnol. 1998. PMID: 9702770 Review.
Cited by
-
Acetamide hydrolyzing activity of Bacillus megaterium F-8 with bioremediation potential: optimization of production and reaction conditions.Environ Sci Pollut Res Int. 2014;21(14):8822-30. doi: 10.1007/s11356-014-2818-7. Epub 2014 Apr 11. Environ Sci Pollut Res Int. 2014. PMID: 24723348
-
Advances in cloning, structural and bioremediation aspects of nitrile hydratases.Mol Biol Rep. 2019 Aug;46(4):4661-4673. doi: 10.1007/s11033-019-04811-w. Epub 2019 Jun 14. Mol Biol Rep. 2019. PMID: 31201677 Review.
-
Structure-Based Engineering of Amidase from Pantoea sp. for Efficient 2-Chloronicotinic Acid Biosynthesis.Appl Environ Microbiol. 2019 Feb 20;85(5):e02471-18. doi: 10.1128/AEM.02471-18. Print 2019 Mar 1. Appl Environ Microbiol. 2019. PMID: 30578259 Free PMC article.
-
Characterization of acetonitrile-tolerant marine bacterium Exiguobacterium sp. SBH81 and its tolerance mechanism.Microbes Environ. 2012;27(1):30-5. doi: 10.1264/jsme2.me11228. Epub 2011 Oct 5. Microbes Environ. 2012. PMID: 21971080 Free PMC article.
-
Enantioselective hydrolysis of (R)-2, 2-dimethylcyclopropane carboxamide by immobilized cells of an R-amidase-producing bacterium, Delftia tsuruhatensis CCTCC M 205114, on an alginate capsule carrier.J Ind Microbiol Biotechnol. 2010 May;37(5):503-10. doi: 10.1007/s10295-010-0696-7. Epub 2010 Feb 23. J Ind Microbiol Biotechnol. 2010. PMID: 20177734
References
-
- Asano, Y., and Y. Kato. 1998. Z-phenylacetaldoxime degradation by a novel aldoxime dehydratase from Bacillus sp. strain OxB-1. FEMS Microbiol. Lett. 158:185-190.
-
- Beacham, I. R. 1987. Silent genes in prokaryotes. FEMS Microbiol. Rev. 46:409-417.
-
- Bigey, F. 1995. Ph.D. thesis. Ecole Nationale Superieure Agronomique de Montpellier, Montpellier, France.
-
- Bisset, K. A., and F. Moore. 1950. Jensenia, a new genus of the Actinomycetales. J. Gen. Microbiol. 4:280. - PubMed
-
- Blakey, A. J. 1994. Isolation of nitrile utilising microorganisms and physiological and biochemical investigation of Rhodococcus nov. sp. AJ270 and investigation of its nitrile hydratase. Ph.D. thesis. University of Sunderland, Sunderland, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

