Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10
- PMID: 14532028
- PMCID: PMC201213
- DOI: 10.1128/AEM.69.10.5802-5811.2003
Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10
Abstract
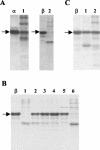
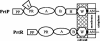
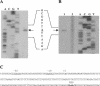
A novel proteinase, PrtR, produced by the human vaginal isolate Lactobacillus rhamnosus strain BGT10 was identified and genetically characterized. The prtR gene and flanking regions were cloned and sequenced. The deduced amino acid sequence of PrtR shares characteristics that are common for other cell envelope proteinases (CEPs) characterized to date, but in contrast to the other cell surface subtilisin-like serine proteinases, it has a smaller and somewhat different B domain and lacks the helix domain, and the anchor domain has a rare sorting signal sequence. Furthermore, PrtR lacks the insert domain, which otherwise is situated inside the catalytic serine protease domain of all CEPs, and has a different cell wall spacer (W) domain similar to that of the cell surface antigen I and II polypeptides expressed by oral and vaginal streptococci. Moreover, the PrtR W domain exhibits significant sequence homology to the consensus sequence that has been shown to be the hallmark of human intestinal mucin protein. According to its alpha(S1)- and beta-casein cleavage efficacy, PrtR is an efficient proteinase at pH 6.5 and is distributed throughout all L. rhamnosus strains tested. Proteinase extracts of the BGT10 strain obtained with Ca(2+)-free buffer at pH 6.5 were proteolytically active. The prtR promoter-like sequence was determined, and the minimal promoter region was defined by use of prtR-gusA operon fusions. The prtR expression is Casitone dependent, emphasizing that nitrogen depletion elevates its transcription. This is in correlation with the catalytic activity of the PrtR proteinase.
Figures

Similar articles
-
Analysis of the presence of prtR proteinase gene in natural isolates of Lactobacillus rhamnosus.Folia Microbiol (Praha). 2006;51(6):535-40. doi: 10.1007/BF02931617. Folia Microbiol (Praha). 2006. PMID: 17455789
-
The presence of prtP proteinase gene in natural isolate Lactobacillus plantarum BGSJ3-18.Lett Appl Microbiol. 2010 Jan;50(1):43-9. doi: 10.1111/j.1472-765X.2009.02748.x. Lett Appl Microbiol. 2010. PMID: 19843212
-
Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes.Microbiology (Reading). 1998 Jun;144 ( Pt 6):1583-1892. doi: 10.1099/00221287-144-6-1583. Microbiology (Reading). 1998. PMID: 9639929
-
Multi-domain, cell-envelope proteinases of lactic acid bacteria.Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):139-55. Antonie Van Leeuwenhoek. 1999. PMID: 10532376 Review.
-
Original features of cell-envelope proteinases of Lactobacillus helveticus. A review.Int J Food Microbiol. 2011 Mar 15;146(1):1-13. doi: 10.1016/j.ijfoodmicro.2011.01.039. Epub 2011 Feb 2. Int J Food Microbiol. 2011. PMID: 21354644 Review.
Cited by
-
Transcriptome analysis of probiotic Lactobacillus casei Zhang during fermentation in soymilk.J Ind Microbiol Biotechnol. 2012 Jan;39(1):191-206. doi: 10.1007/s10295-011-1015-7. Epub 2011 Jul 22. J Ind Microbiol Biotechnol. 2012. PMID: 21779970
-
Identification of Antihypertensive Peptides Derived from Low Molecular Weight Casein Hydrolysates Generated during Fermentation by Bifidobacterium longum KACC 91563.Korean J Food Sci Anim Resour. 2015;35(6):738-47. doi: 10.5851/kosfa.2015.35.6.738. Epub 2015 Dec 31. Korean J Food Sci Anim Resour. 2015. PMID: 26877633 Free PMC article.
-
Current Perspectives on Antihypertensive Probiotics.Probiotics Antimicrob Proteins. 2017 Jun;9(2):91-101. doi: 10.1007/s12602-016-9241-y. Probiotics Antimicrob Proteins. 2017. PMID: 27900619 Review.
-
Brazilian Kefir-Fermented Sheep's Milk, a Source of Antimicrobial and Antioxidant Peptides.Probiotics Antimicrob Proteins. 2018 Sep;10(3):446-455. doi: 10.1007/s12602-017-9365-8. Probiotics Antimicrob Proteins. 2018. PMID: 29285743
-
Phylogenetic survey of the subtilase family and a data-mining-based search for new subtilisins from Bacillaceae.Front Microbiol. 2022 Sep 26;13:1017978. doi: 10.3389/fmicb.2022.1017978. eCollection 2022. Front Microbiol. 2022. PMID: 36225363 Free PMC article.
References
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 12:248-254. - PubMed
-
- Bruinenberg, P. G., P. Doesburg, A. C. Alting, F. A. Exterkate, W. M. de Vos, and R. J. Siezen. 1994. Evidence for a large dispensable segment in the subtilisin-like catalytic domain of the Lactococcus lactis cell-envelope proteinase. Protein Eng. 7:991-996. - PubMed
-
- Chassy, B. M., and L. Flickinger. 1987. Transformation of Lactobacillus casei by electroporation. FEMS Microbiol. Lett. 44:173-177.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

