Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae
- PMID: 14532041
- PMCID: PMC201209
- DOI: 10.1128/AEM.69.10.5892-5897.2003
Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae
Abstract
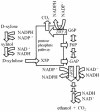
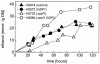
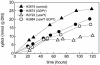
Pentose fermentation to ethanol with recombinant Saccharomyces cerevisiae is slow and has a low yield. A likely reason for this is that the catabolism of the pentoses D-xylose and L-arabinose through the corresponding fungal pathways creates an imbalance of redox cofactors. The process, although redox neutral, requires NADPH and NAD+, which have to be regenerated in separate processes. NADPH is normally generated through the oxidative part of the pentose phosphate pathway by the action of glucose-6-phosphate dehydrogenase (ZWF1). To facilitate NADPH regeneration, we expressed the recently discovered gene GDP1, which codes for a fungal NADP+-dependent D-glyceraldehyde-3-phosphate dehydrogenase (NADP-GAPDH) (EC 1.2.1.13), in an S. cerevisiae strain with the D-xylose pathway. NADPH regeneration through an NADP-GAPDH is not linked to CO2 production. The resulting strain fermented D-xylose to ethanol with a higher rate and yield than the corresponding strain without GDP1; i.e., the levels of the unwanted side products xylitol and CO2 were lowered. The oxidative part of the pentose phosphate pathway is the main natural path for NADPH regeneration. However, use of this pathway causes wasteful CO2 production and creates a redox imbalance on the path of anaerobic pentose fermentation to ethanol because it does not regenerate NAD+. The deletion of the gene ZWF1 (which codes for glucose-6-phosphate dehydrogenase), in combination with overexpression of GDP1 further stimulated D-xylose fermentation with respect to rate and yield. Through genetic engineering of the redox reactions, the yeast strain was converted from a strain that produced mainly xylitol and CO2 from D-xylose to a strain that produced mainly ethanol under anaerobic conditions.
Figures
References
-
- Alexander, M. A., V. W. Yang, and T. W. Jeffries. 1988. Levels of pentose phosphate pathway enzymes from Candida shehatae grown in continuous culture. Appl. Microbiol. Biotechnol. 29:282-288.
-
- Aristidou, A., P. Richard, L. Ruohonen, M. Toivari, J. Londesborough, and M. Penttilä. 1999. Redox balance in fermenting yeast. Monogr. Eur. Brewery Convent. 28:161-169.
-
- Boles, E., H. W. Gohlmann, and F. K. Zimmermann. 1996. Cloning of a second gene encoding 5-phosphofructo-2-kinase in yeast, and characterization of mutant strains without fructose-2, 6-bisphosphate. Mol. Microbiol. 20:65-76. - PubMed
-
- Boles, E., W. Lehnert, and F. K. Zimmermann. 1993. The role of the NAD-dependent glutamate dehydrogenase in restoring growth on glucose of a Saccharomyces cerevisiae phosphoglucose isomerase mutant. Eur. J. Biochem. 217:469-477. - PubMed
-
- Bruinenberg, P. M., R. Jonker, J. P. van Dijken, and W. A. Scheffers. 1985. Utilization of formate as an additional energy source by glucose-limited chemostat cultures of Candida utilis CBS621 and Saccharomyces cerevisiae CBS 8066. Evidence for the absence of transhydrogenase activity in yeasts. Arch. Microbiol. 142:302-306.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

