Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1
- PMID: 14532048
- PMCID: PMC201243
- DOI: 10.1128/AEM.69.10.5941-5949.2003
Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1
Abstract
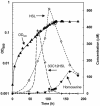
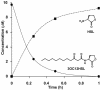
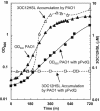
Acyl-homoserine lactones (AHLs) are employed by several Proteobacteria as quorum-sensing signals. Past studies have established that these compounds are subject to biochemical decay and can be used as growth nutrients. Here we describe the isolation of a soil bacterium, Pseudomonas strain PAI-A, that degrades 3-oxododecanoyl-homoserine lactone (3OC12HSL) and other long-acyl, but not short-acyl, AHLs as sole energy sources for growth. The small-subunit rRNA gene from strain PAI-A was 98.4% identical to that of Pseudomonas aeruginosa, but the soil isolate did not produce obvious pigments or AHLs or grow under denitrifying conditions or at 42 degrees C. The quorum-sensing bacterium P. aeruginosa, which produces both 3OC12HSL and C4HSL, was examined for the ability to utilize AHLs for growth. It did so with a specificity similar to that of strain PAI-A, i.e., degrading long-acyl but not short-acyl AHLs. In contrast to the growth observed with strain PAI-A, P. aeruginosa strain PAO1 growth on AHLs commenced only after extremely long lag phases. Liquid-chromatography-atmospheric pressure chemical ionization-mass spectrometry analyses indicate that strain PAO1 degrades long-acyl AHLs via an AHL acylase and a homoserine-generating HSL lactonase. A P. aeruginosa gene, pvdQ (PA2385), has previously been identified as being a homologue of the AHL acylase described as occurring in a Ralstonia species. Escherichia coli expressing pvdQ catalyzed the rapid inactivation of long-acyl AHLs and the release of HSL. P. aeruginosa engineered to constitutively express pvdQ did not accumulate its 3OC12HSL quorum signal when grown in rich media. However, pvdQ knockout mutants of P. aeruginosa were still able to grow by utilizing 3OC12HSL. To our knowledge, this is the first report of the degradation of AHLs by pseudomonads or other gamma-Proteobacteria, of AHL acylase activity in a quorum-sensing bacterium, of HSL lactonase activity in any bacterium, and of AHL degradation with specificity only towards AHLs with long side chains.
Figures


References
-
- Anzai, Y., H. Kim, J. Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50:1563-1589. - PubMed
-
- Anzai, Y., Y. Kudo, and H. Oyaizu. 1997. The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int. J. Syst. Bacteriol. 47:249-251. - PubMed
-
- Bennasar, A., R. Rossello-Mora, J. Lalucat, and E. R. Moore. 1996. 16S rRNA gene sequence analysis relative to genomovars of Pseudomonas stutzeri and proposal of Pseudomonas balearica sp. nov. Int. J. Syst. Bacteriol. 46:200-205. - PubMed
-
- Cronin, C. N., and W. S. McIntire. 1999. pUCP-Nco and pUCP-Nde: Escherichia-Pseudomonas shuttle vectors for recombinant protein expression in Pseudomonas. Anal. Biochem. 272:112-115. - PubMed
-
- Domenech Fernandez-Garayzabal, J. F., P. Lawson, J. A. Garcia, M. T. Cutuli, M. Blanco, A. Gibello, M. A. Moreno, M. D. Collins, and L. Dominguez. 1997. Winter disease outbreak in sea-bream (Sparus aurata) associated with Pseudomonas anguilliseptica infection. Aquaculture 156:317-326.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

