Detection of Prochlorothrix in brackish waters by specific amplification of pcb genes
- PMID: 14532086
- PMCID: PMC201227
- DOI: 10.1128/AEM.69.10.6243-6249.2003
Detection of Prochlorothrix in brackish waters by specific amplification of pcb genes
Abstract
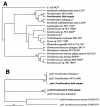
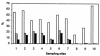
Prochlorothrix hollandica is the only filamentous chlorophyll b (Chlb)-containing oxyphotobacterium that has been found in freshwater habitats to date. Chlb serves as a light-harvesting pigment which is bound to special binding proteins (Pcb). Even though Prochlorothrix was initially characterized as a highly salt-sensitive species, we detected it in a brackish water environment that is characterized by salinities of up to 12 practical salinity units. Using PCR and reverse transcription, we amplified pcb gene fragments of phytoplankton samples taken along a salinity gradient in the eutrophic Darss-Zingst estuary (southern Baltic Sea). After sequencing, high levels of homology to the pcbB and pcbC genes of P. hollandica were found. Furthermore, autofluorescence of Prochlorothrix-like filaments that indicated that Chlb was present was detected in enrichment cultures prepared from the estuarine phytoplankton. The detection of Chlb-containing filaments, as well as the pcb and 16S ribosomal DNA sequences, suggests that Prochlorothrix is an indigenous genus in the Darss-Zingst estuary and may also inhabit many other brackish water environments. The potential of using pcb gene detection to differentiate Prochlorothrix from morphologically indistinguishable species belonging to the genera Pseudanabaena and Planktothrix (Oscillatoria) in phytoplankton analyses is discussed.
Figures



Similar articles
-
Salinity tolerance of the chlorophyll b-synthesizing cyanobacterium Prochlorothrix hollandica strain SAG 10.89.Microb Ecol. 2008 May;55(4):685-96. doi: 10.1007/s00248-007-9311-5. Microb Ecol. 2008. PMID: 17874260
-
Transcript analysis of the pcbABC genes encoding the antenna apoproteins in the photosynthetic prokaryote, Prochlorothrix hollandica.FEMS Microbiol Lett. 1998 Nov 15;168(2):187-94. doi: 10.1111/j.1574-6968.1998.tb13272.x. FEMS Microbiol Lett. 1998. PMID: 9835028
-
The 38 kDa chlorophyll a/b protein of the prokaryote Prochlorothrix hollandica is encoded by a divergent pcb gene.Plant Mol Biol. 1998 Mar;36(5):709-16. doi: 10.1023/a:1005930210515. Plant Mol Biol. 1998. PMID: 9526503
-
Localization of Pcb antenna complexes in the photosynthetic prokaryote Prochlorothrix hollandica.Biochim Biophys Acta. 2010 Jan;1797(1):89-97. doi: 10.1016/j.bbabio.2009.09.002. Epub 2009 Sep 15. Biochim Biophys Acta. 2010. PMID: 19761753
-
The prochlorophytes: are they more than just chlorophyll a/b-containing cyanobacteria?Crit Rev Microbiol. 1993;19(1):43-59. doi: 10.3109/10408419309113522. Crit Rev Microbiol. 1993. PMID: 8481212 Review.
Cited by
-
Salinity tolerance of the chlorophyll b-synthesizing cyanobacterium Prochlorothrix hollandica strain SAG 10.89.Microb Ecol. 2008 May;55(4):685-96. doi: 10.1007/s00248-007-9311-5. Microb Ecol. 2008. PMID: 17874260
-
Blooms of single bacterial species in a coastal lagoon of the southwestern Atlantic Ocean.Appl Environ Microbiol. 2006 Oct;72(10):6560-8. doi: 10.1128/AEM.01089-06. Appl Environ Microbiol. 2006. PMID: 17021206 Free PMC article.
-
Cyanobacteria of the genus prochlorothrix.Front Microbiol. 2012 May 21;3:173. doi: 10.3389/fmicb.2012.00173. eCollection 2012. Front Microbiol. 2012. PMID: 22783229 Free PMC article.
References
-
- Barker, G. L., B. A. Handley, P. Vacharapiyasophon, J. R. Stevens, and P. K. Hayes. 2000. Allele-specific PCR shows that genetic exchange occurs among genetically diverse Nodularia (cyanobacteria) filaments in the Baltic Sea. Microbiology 146:2865-2875. - PubMed
-
- Bullerjahn, G. S., and A. F. Post. 1993. The prochlorophytes: are they more than just chlorophyll a/b-containing cyanobacteria? Crit. Rev. Microbiol. 19:43-59. - PubMed
-
- Burger-Wiersma, T., M. Veenhuis, H. J. Korthals, C. C. M. Van de Wiel, and L. R. Mur. 1986. A new prokaryote containing chlorophylls a and b. Nature 320:262-264.
-
- Burger-Wiersma, T., L. J. Stal, and L. M. Mur. 1989. Prochlorothrix hollandica gen. nov., sp. nov., a filamentous oxygenic photoautotrophic prokaryote containing chlorophylls a and b: assignment to Prochlorotrichaceae fam. nov. and order Prochlorales Florenzano, Balloni, and Materassi 1986, with emendation of the ordinal description. Int. J. Syst. Bacteriol. 39:250-257.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

