Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci
- PMID: 14532213
- PMCID: PMC254362
- DOI: 10.1128/JCM.41.10.4740-4744.2003
Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci
Abstract
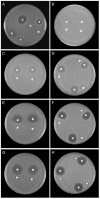
Resistance to macrolides in staphylococci may be due to active efflux (encoded by msrA) or ribosomal target modification (macrolide-lincosamide-streptogramin B [MLSB] resistance; usually encoded by ermA or ermC). MLSB resistance is either constitutive or inducible following exposure to a macrolide. Induction tests utilize closely approximated erythromycin and clindamycin disks; the flattening of the clindamycin zone adjacent to the erythromycin disk indicates inducible MLSB resistance. The present study reassessed the reliability of placing erythromycin and clindamycin disks in adjacent positions (26 to 28 mm apart) in a standard disk dispenser, compared to distances of 15 or 20 mm. A group of 130 clinical isolates of Staphylococcus aureus and 100 isolates of erythromycin-resistant coagulase-negative staphylococci (CNS) were examined by disk approximation; all CNS isolates and a subset of S. aureus isolates were examined by PCR for ermA, ermC, and msrA. Of 114 erythromycin-resistant S. aureus isolates, 39 demonstrated constitutive resistance to clindamycin, while 33 showed inducible resistance by disk approximation at all three distances. Only one isolate failed to clearly demonstrate induction at 26 mm. Of 82 erythromycin-resistant CNS isolates that contained ermA or ermC, 57 demonstrated constitutive clindamycin resistance, and 25 demonstrated inducible resistance, at 20 and 26 mm. None of the 42 S. aureus isolates or 18 CNS isolates containing only msrA and none of the erythromycin-susceptible isolates yielded positive disk approximation tests. Simple placement of erythromycin and clindamycin disks at a distance achieved with a standard disk dispenser allowed detection of 97% of S. aureus strains and 100% of CNS strains with inducible MLSB resistance in this study.
Figures
References
-
- Catchpole, I., and K. G. H. Dyke. 1990. A Staphylococcus aureus plasmid that specifies constitutive macrolide-lincosamide-streptogramin B resistance contains a novel deletion in the ermC attenuator. FEMS Microbiol. Lett. 69:43-48. - PubMed
-
- Di Modugno, V., M. Guerrini, S. Shah, and J. Hamilton-Miller. 2002. Low level resistance to oleandomycin as a marker of ermA in staphylococci. J. Antimicrob. Chemother. 49:423-430. - PubMed
-
- Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315-316. - PubMed
-
- Eady, E. A., J. I. Ross, J. L. Tipper, C. E. Walters, J. H. Cove, and W. C. Noble. 1993. Distribution of genes encoding erythromycin ribosomal methylases and an erythromycin efflux pump in epidemiologically distinct groups of staphylococci. J. Antimicrob. Chemother. 31:211-217. - PubMed
-
- Frank, A. L., J. F. Marcinak, P. D. Mahgat, J. T. Tjhio, S. Kelkar, P. C. Schreckenberger, and J. P. Quinn. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21:530-534. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

